8FLE
 
 | |
6PZ8
 
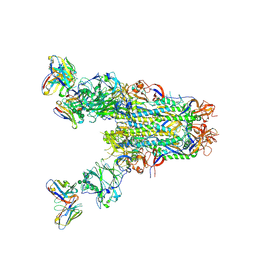 | | MERS S0 trimer in complex with variable domain of antibody G2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G2 heavy chain, ... | | Authors: | Bowman, C.A, Pallesen, J, Ward, A.B. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
6Q0F
 
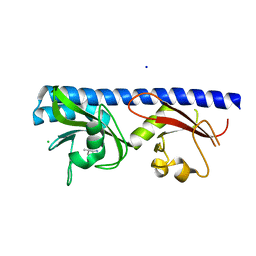 | | Crystal structure of ligand-binding domain of Pseudomonas fluorescens chemoreceptor CtaA in complex with L-valine | | Descriptor: | CHLORIDE ION, Putative methyl-accepting chemotaxis protein, SODIUM ION, ... | | Authors: | Ud-Din, I.A, Khan, M.F, Roujeinikova, A. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Broad Specificity of Amino Acid Chemoreceptor CtaA ofPseudomonas fluorescensIs Afforded by Plasticity of Its Amphipathic Ligand-Binding Pocket.
Mol.Plant Microbe Interact., 33, 2020
|
|
8FL3
 
 | |
8FZV
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Ala-Ala linker variant, expressed with SUMO tag | | Descriptor: | MAGNESIUM ION, Transcription factor ETV6,Anthrax toxin receptor 2, UNKNOWN ATOM OR ION | | Authors: | Pedroza Romo, M.J, Soleimani, S, Doukov, T, Lebedev, A, Moody, J.D. | | Deposit date: | 2023-01-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8FLB
 
 | |
8FLF
 
 | |
6Q30
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 5 | | Descriptor: | (7-carboxy-1-benzothiophen-2-yl)-tris(oxidanyl)boranuide, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
8D9E
 
 | | gRAMP-match PFS target | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*UP*A)-3'), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
7BJT
 
 | | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT | | Descriptor: | Alginate lyase, family PL17, CALCIUM ION, ... | | Authors: | Czjzek, M, Roret, T, Jouanneau, D, Le Duff, N, Jeudy, A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT.
Glycobiology, 31, 2021
|
|
7QTG
 
 | | Crystal Structure of the Fe(II)/alpha-ketoglutarate dependent dioxygenase PlaO1 | | Descriptor: | FE (II) ION, PlaO1, SULFATE ION | | Authors: | Lukat, P, Daum, M, Bechthold, A, Einsle, O. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Structural investigations on the Fe(II)/alpha-ketoglutarate dependent dioxygense PlaO1 from Streptomyces sp. Tu6071
Thesis, 2011
|
|
8SPK
 
 | | Crystal structure of Antarctic PET-degrading enzyme | | Descriptor: | Lipase 1, MALONATE ION | | Authors: | Furtado, A.A, Blazquez-Sanchez, P, Grinen, A, Vargas, J.A, Leonardo, D.A, Sculaccio, S.A, Pereira, H.M, Diez, B, Garratt, R.C, Ramirez-Sarmiento, C.A. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the catalytic activity of an Antarctic PET-degrading enzyme by loop exchange.
Protein Sci., 32, 2023
|
|
6Q4W
 
 | | Structure of MPT-1, a GDP-Man-dependent mannosyltransferase from Leishmania mexicana | | Descriptor: | LmxM MPT-1 | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
7QSJ
 
 | | Methylmannose polysaccharide hydrolase MmpH from M. hassiacum | | Descriptor: | GLYCEROL, MAGNESIUM ION, Methylmannose polysaccharide hydrolase (MmpH), ... | | Authors: | Ripoll-Rozada, J, Manso, J.A, Pereira, P.J.B. | | Deposit date: | 2022-01-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Self-recycling and partially conservative replication of mycobacterial methylmannose polysaccharides.
Commun Biol, 6, 2023
|
|
6Q73
 
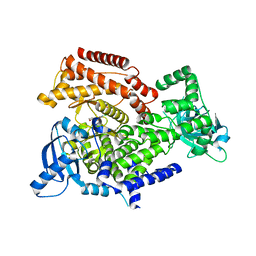 | | PI3K delta in complex with N[2chloro5(3,6dihydro2Hpyran4yl)pyridin3yl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[2-chloranyl-5-(3,6-dihydro-2~{H}-pyran-4-yl)pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Rowland, P, Down, K, Barton, N. | | Deposit date: | 2018-12-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of Potent, Efficient, and Selective Inhibitors of Phosphoinositide 3-Kinase delta through a Deconstruction and Regrowth Approach.
J.Med.Chem., 61, 2018
|
|
8D9G
 
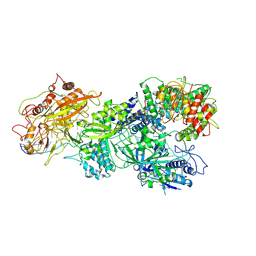 | | gRAMP-TPR-CHAT Non match PFS target RNA(Craspase) | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (36-MER), ... | | Authors: | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
6QEO
 
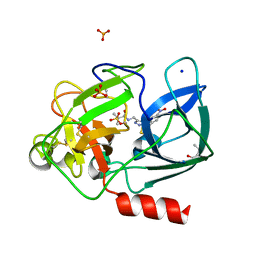 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the 3-Oxo-beta-Sultam inhibitor LMC269 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-methyl-1-[[1-[(4-nitrophenyl)methyl]-1,2,3-triazol-4-yl]methylamino]-1-oxidanylidene-propane-2-sulfonic acid, Chymotrypsin-like elastase family member 1, ... | | Authors: | Brito, J.A, Almeida, V.T, Carvalho, L.M, Moreira, R, Archer, M. | | Deposit date: | 2019-01-08 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 3-Oxo-beta-sultam as a Sulfonylating Chemotype for Inhibition of Serine Hydrolases and Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
6QGD
 
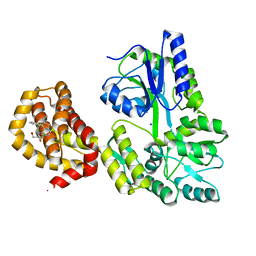 | | Structure of human Mcl-1 in complex with thienopyrimidine inhibitor | | Descriptor: | 2-[(6-ethyl-5-phenyl-thieno[2,3-d]pyrimidin-4-yl)amino]-3-oxidanyl-propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
8FA2
 
 | |
8FA1
 
 | |
7QUI
 
 | | Crystal structure of the N-terminal domain of Siglec-8 in complex with sulfonamide sialoside analogue | | Descriptor: | (2~{S},4~{S},5~{R},6~{R})-5-acetamido-2-[(2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-acetamido-2-(hydroxymethyl)-4,6-bis(oxidanyl)oxan-3-yl]oxy-3,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-4-yl]oxy-6-[(1~{R},2~{R})-3-(naphthalen-2-ylsulfonylamino)-1,2-bis(oxidanyl)propyl]-4-oxidanyl-oxane-2-carboxylic acid, Sialic acid-binding Ig-like lectin 8 | | Authors: | Lenza, M.P, Oyenarte, I, Atxabal, U, Nycholat, C, Franconetti, A, Quintana, J.I, Delgado, S, Unione, L, Paulson, J, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.352 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
7QU6
 
 | | Crystal structure of the N-terminal domain of Siglec-8 | | Descriptor: | Sialic acid-binding Ig-like lectin 8 | | Authors: | Lenza, M.P, Atxabal, U, Nycholat, C.M, Oyenarte, I, Paulson, J.C, Franconetti, A, Quintana, J.I, Unione, L, Delgado, S, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
8DFY
 
 | | Crystal structure of Human BTN2A1 Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, ... | | Authors: | Fulford, T.S, Soliman, C, Castle, R.G, Rigau, M, Ruan, Z, Dolezal, O, Seneviratna, R, Brown, H.G, Hanssen, E, Hammet, A, Li, S, Redmond, S.J, Chung, A, Gorman, M.A, Parker, M.W, Patel, O, Peat, T.S, Newman, J, Behren, A, Gherardin, N.A, Godfrey, D.I, Uldrich, A.P. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.552 Å) | | Cite: | Vgamma9-Vdelta2 T cells recognize butyrophilin 2A1 and 3A1 heteromers
To Be Published
|
|
6QJN
 
 | |
7MDJ
 
 | | The structure of KcsA in complex with a synthetic Fab | | Descriptor: | Fab heavy chain, Fab light chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Slezak, T, Blackowicz, L, Kossiakoff, A, Roux, B. | | Deposit date: | 2021-04-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Engineering of a synthetic antibody fragment for structural and functional studies of K+ channels.
J.Gen.Physiol., 154, 2022
|
|
