7O6S
 
 | | Crystal structure of a shortened IpgC variant in complex with N-(2H-1,3-benzodioxol-5-ylmethyl)cyclopentanamine | | Descriptor: | CHLORIDE ION, Chaperone protein IpgC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gardonyi, M, Heine, A, Klebe, G. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a shortened IpgC variant in complex with N-(2H-1,3-benzodioxol-5-ylmethyl)cyclopentanamine
To be published
|
|
7OD5
 
 | | F(M197)H mutant structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV LSP crystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of the Rhodobacter sphaeroides reaction center with an M197 Phe→His substitution clarifies the properties of the mutant complex.
Iucrj, 9, 2022
|
|
7OB6
 
 | |
8DES
 
 | | Gokushovirus EC6098 | | Descriptor: | Major capsid protein, Putative DNA binding protein | | Authors: | Lee, H, Fane, B.A, Hafenstein, S.L. | | Deposit date: | 2022-06-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM Structure of Gokushovirus Phi EC6098 Reveals a Novel Capsid Architecture for a Single-Scaffolding Protein, Microvirus Assembly System.
J.Virol., 96, 2022
|
|
7QD8
 
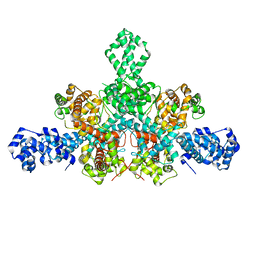 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in apo state | | Descriptor: | Transposase for transposon Tn4430 | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
7QD4
 
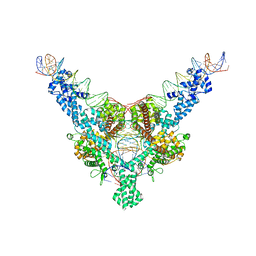 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with 100 bp long transposon end DNA | | Descriptor: | IR100 DNA substrate, none transferred strand, transferred strand, ... | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
6PY1
 
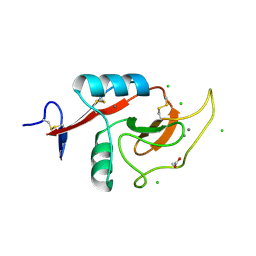 | | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, ACETATE ION, C-type lectin domain family 10 member A, ... | | Authors: | Birrane, G, Murphy, P.V, Gabba, A, Luz, J.G. | | Deposit date: | 2019-07-28 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to GalNAc and the Tumor-Associated Tn Antigen.
Biochemistry, 60, 2021
|
|
6NB5
 
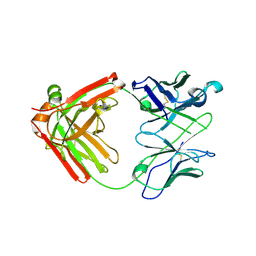 | | Crystal structure of anti- MERS-CoV human neutralizing LCA60 antibody Fab fragment | | Descriptor: | LCA60 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6Q0G
 
 | |
6Q0R
 
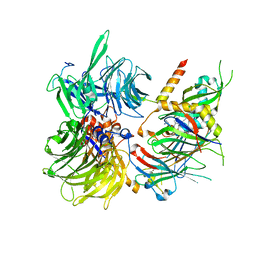 | | Structure of DDB1-DDA1-DCAF15 complex bound to E7820 and RBM39 | | Descriptor: | 3-cyano-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
8CVL
 
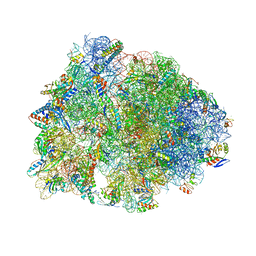 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-NH-tRNAphe, peptidyl P-site fMTHSMRC-NH-tRNAmet, and deacylated E-site tRNAphe at 2.30A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-05-18 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes.
Nat.Chem., 15, 2023
|
|
7AAT
 
 | |
7QD6
 
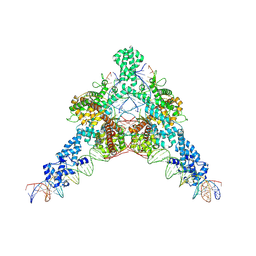 | | Cryo-EM structure of Tn4430 TnpA transposase from Tn3 family in complex with strand-transfer like DNA product | | Descriptor: | IR71st non transferred strand, IR71st transferred strand, Transposase for transposon Tn4430 | | Authors: | Shkumatov, A.V, Oger, C.A, Aryanpour, N, Hallet, B.F, Efremov, R.G. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insight into Tn3 family transposition mechanism.
Nat Commun, 13, 2022
|
|
6WPT
 
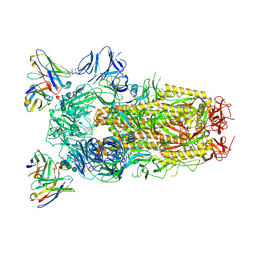 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
5M11
 
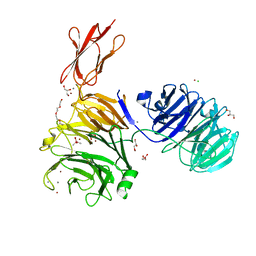 | | Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lasica, A.M, Goulas, T, Mizgalska, D, Zhou, X, Ksiazek, M, Madej, M, Guo, Y, Guevara, T, Nowak, M, Potempa, B, Goel, A, Sztukowska, M, Prabhakar, A.T, Bzowska, M, Widziolek, M, Thogersen, I.B, Enghild, J.J, Simonian, M, Kulczyk, A.W, Nguyen, K.-A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis.
Sci Rep, 6, 2016
|
|
6ZN0
 
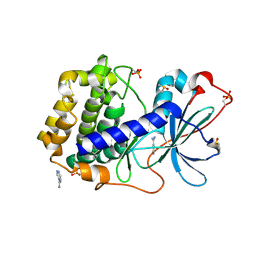 | | Crystal structure of cAMP-dependent protein kinase A (CHO PKA) in complex with isonicotinamidine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DIMETHYL SULFOXIDE, ISONICOTINAMIDINE, ... | | Authors: | Oebbeke, M, Heine, A, Klebe, G. | | Deposit date: | 2020-07-06 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6NKD
 
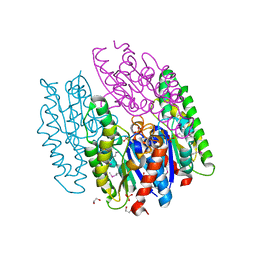 | | Crystal Structure of the Lipase Lip_vut3 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut3 from Goat Rumen metagenome.
To Be Published
|
|
6PUM
 
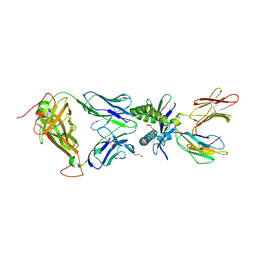 | | Structure of human MAIT A-F7 TCR in complex with human MR1-2'D-5-OP-RU | | Descriptor: | 1,2-dideoxy-1-({2,6-dioxo-5-[(E)-(2-oxopropylidene)amino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-erythro-pentitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Awad, W, Keller, A.N, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
6XZB
 
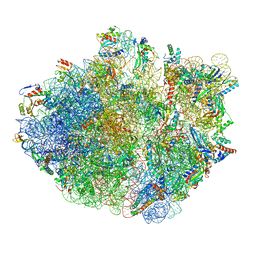 | | E. coli 70S ribosome in complex with dirithromycin, fMet-Phe-tRNA(Phe) and deacylated tRNA(iMet) (focused classification). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pichkur, E.B, Polikanov, Y.S, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2020-02-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to the E. coli 70S ribosome.
Rna, 26, 2020
|
|
6SPC
 
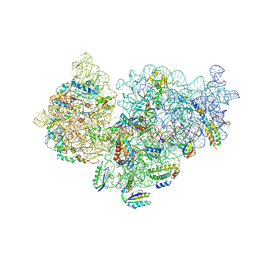 | | Pseudomonas aeruginosa 30s ribosome from an aminoglycoside resistant clinical isolate | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7OIW
 
 | |
8FG1
 
 | | Human diaphanous inhibitory domain bound to diaphanous autoregulatory domain | | Descriptor: | Protein diaphanous homolog 1 | | Authors: | Ramirez, L.M.S, Theophall, G, Premo, A, Manigrasso, M, Yepuri, G, Burz, D, Ramasamy, R, Schmidt, A.M, Shekhtman, A. | | Deposit date: | 2022-12-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of the productive encounter complex results in dysregulation of DIAPH1 activity.
J.Biol.Chem., 299, 2023
|
|
6NSY
 
 | | X-ray reduced Catalase 3 From N.Crassa in Cpd I state (0.263 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
6PUL
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1 3'D-5-OP-RU | | Descriptor: | 1,3-dideoxy-1-({2,6-dioxo-5-[(E)-(2-oxopropylidene)amino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-erythro-pentitol, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Awad, W, Keller, A.N, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
6PW6
 
 | | The HIV-1 Envelope Glycoprotein Clone BG505 SOSIP.664 in Complex with Three Copies of the Bovine Broadly Neutralizing Antibody, NC-Cow1, Fragment Antigen Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Broadly Neutralizing Antibody NC-Cow1 Heavy Chain, ... | | Authors: | Berndsen, Z.T, Ward, A.B. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of broad HIV neutralization by a vaccine-induced cow antibody.
Sci Adv, 6, 2020
|
|
