4UCU
 
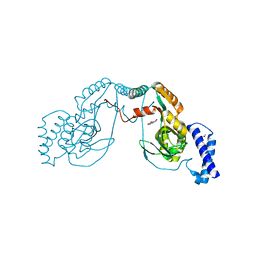 | | Fragment bound to H.influenza NAD dependent DNA ligase | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxyquinoline-2-carboxylic acid, DNA LIGASE | | 著者 | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UMR
 
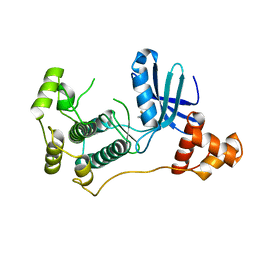 | | Structure of MELK in complex with inhibitors | | 分子名称: | 4-fluoro-N-(1,2,3,4-tetrahydroisoquinolin-7-yl)benzamide, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | 著者 | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | 登録日 | 2014-05-20 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UCS
 
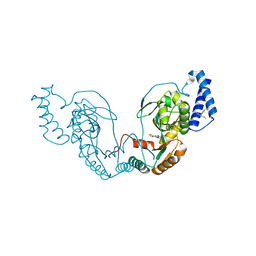 | | Fragment bound to H.influenza NAD dependent DNA ligase | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5-amino-3-(furan-2-yl)-1H-1,2,4-triazole-1-carboxamide, DNA LIGASE | | 著者 | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UHJ
 
 | |
4UCO
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 7-amino-2-tert-butyl-4-(1H-pyrrol-2-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, DNA LIGASE | | 著者 | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
6RU4
 
 | |
4UMO
 
 | |
6NJF
 
 | | Solution NMR Structure of DANCER3-F34A, a rigid and natively folded single mutant of the dynamic protein DANCER-3 | | 分子名称: | Immunoglobulin G-binding protein G | | 著者 | Damry, A.M, Mayer, M.M, Goto, N.K, Chica, R.A. | | 登録日 | 2019-01-03 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Origin of conformational dynamics in a globular protein.
Commun Biol, 2, 2019
|
|
4UCV
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-methoxy-2,3-dimethylquinoxalin-5-ol, DNA LIGASE | | 著者 | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UOB
 
 | | Crystal structure of Deinococcus radiodurans Endonuclease III-3 | | 分子名称: | ACETATE ION, COBALT (II) ION, ENDONUCLEASE III-3, ... | | 著者 | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | 登録日 | 2014-06-02 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
6O41
 
 | |
4UCR
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxy-2-methylquinoline-6-carboxamide, DNA LIGASE | | 著者 | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
6RZ6
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2570366 (C2221 space group) | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(2-chloranyl-5-fluoranyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3- dihydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | 著者 | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | 登録日 | 2019-06-12 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
6O4Z
 
 | | Structure of HLA-A2:01 with peptide MM92 | | 分子名称: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | 著者 | Ying, G, Bitra, A, Zajonc, D.M. | | 登録日 | 2019-03-01 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
6O51
 
 | | Structure of HLA-A2:01 with peptide MM90 | | 分子名称: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | 著者 | Ying, G, Bitra, A, Zajonc, D.M. | | 登録日 | 2019-03-01 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Anin silico-in vitroPipeline Identifying an HLA-A*02:01+KRAS G12V+Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients.
Front Immunol, 10, 2019
|
|
8ILU
 
 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | 分子名称: | (2R,3R,4R,5R,6S)-2-(hydroxymethyl)-6-[2-(2-methyl-1,3-benzothiazol-6-yl)-1,2,4-triazol-3-yl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, SODIUM ION, ... | | 著者 | Kumar, A, Jinal, S, Raman, S, Ghosh, K. | | 登録日 | 2023-03-04 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Identification of benzothiazole derived monosaccharides as potent, selective, and orally bioavailable inhibitors of human and mouse galectin-3; a rare example of using a S···O binding interaction for drug design.
Bioorg.Med.Chem., 101, 2024
|
|
8TSY
 
 | |
8TSZ
 
 | |
8TT4
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=6.0 | | 分子名称: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | 著者 | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | 登録日 | 2023-08-12 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
4UUH
 
 | | X-ray crystal structure of human TNKS in complex with a small molecule inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 5-methyl-3-[4-(piperazin-1-ylmethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, ... | | 著者 | Oliver, A.W, Rajasekaran, M.B, Pearl, L.H. | | 登録日 | 2014-07-28 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Design and Discovery of 3-Aryl-5-Substituted-Isoquinolin-1- Ones as Potent and Selective Tankyrase Inhibitors
Medchemcommm, 6, 2015
|
|
8TT0
 
 | |
8TV4
 
 | |
4UV8
 
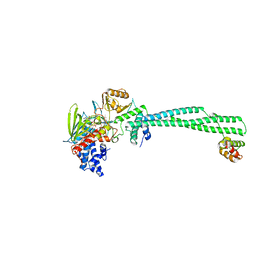 | | LSD1(KDM1A)-CoREST in complex with 1-Benzyl-Tranylcypromine | | 分子名称: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4S)-5-[(4aS,10aS)-4a-[(1S)-3-azanylidene-1,4-diphenyl-butyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-5,10a-dihydro-1H-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentoxy]-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | 登録日 | 2014-08-05 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
8TT2
 
 | | Pseudomonas fluorescens isocyanide hydratase pH=5.4 | | 分子名称: | 1,2-ETHANEDIOL, Isonitrile hydratase InhA | | 著者 | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | 登録日 | 2023-08-12 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Changes in an Enzyme Ensemble During Catalysis Observed by High Resolution XFEL Crystallography.
Biorxiv, 2023
|
|
6CK5
 
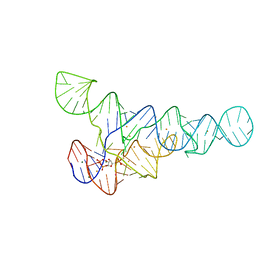 | | PRPP riboswitch from T. mathranii bound to PRPP | | 分子名称: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, BARIUM ION, MAGNESIUM ION, ... | | 著者 | Knappenberger, A.J, Reiss, C.W, Strobel, S.A. | | 登録日 | 2018-02-27 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA.
Elife, 7, 2018
|
|
