6MUH
 
 | | Fluoroacetate dehalogenase, room temperature structure solved by serial 1 degree oscillation crystallography | | 分子名称: | Fluoroacetate dehalogenase | | 著者 | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | 登録日 | 2018-10-23 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
7Q8W
 
 | | Crystal structure of TTBK1 in complex with VNG1.35 (compound 23) | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, Tau-tubulin kinase 1, ... | | 著者 | Chaikuad, A, Nozal, V, Martinez, A, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2021-11-11 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
6PNK
 
 | |
8CWH
 
 | | 200us Temperature-Jump (Dark2) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | 登録日 | 2022-05-19 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
6N59
 
 | | 1.0 Angstrom crystal structure of [FeFe]-hydrogenase | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Zadvornyy, O.A, Keable, S.M, Artz, J.H, Peters, J.W. | | 登録日 | 2018-11-21 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
8CWG
 
 | | 200us Temperature-Jump (Dark1) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | 登録日 | 2022-05-19 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
6PJD
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-32 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-3-methyl-L-valyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | 著者 | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-06-28 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
8CWB
 
 | | Laser Off Temperature-Jump XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | 登録日 | 2022-05-19 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
6PJM
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-35 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3S,5S)-3-hydroxy-5-{[N-(methoxycarbonyl)-3-methyl-L-valyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | 著者 | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-06-28 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
8CWE
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | 登録日 | 2022-05-19 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
7QQK
 
 | | TIR-SAVED effector bound to cA3 | | 分子名称: | RNA (5'-R(P*AP*AP*A)-3'), TIR_SAVED fusion protein | | 著者 | Spagnolo, L, White, M.F, Hogrel, G, Guild, A. | | 登録日 | 2022-01-09 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cyclic nucleotide-induced helical structure activates a TIR immune effector.
Nature, 608, 2022
|
|
6N8B
 
 | | Crystal structure of transcription regulator AcaB from uropathogenic E. coli | | 分子名称: | CALCIUM ION, transcription regulator AcaB | | 著者 | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | 登録日 | 2018-11-29 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
8CWD
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | 登録日 | 2022-05-19 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
6N8A
 
 | | Crystal structure of selenomethionine-containing AcaB from uropathogenic E. coli | | 分子名称: | CHLORIDE ION, transcription regulator AcaB | | 著者 | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | 登録日 | 2018-11-28 | | 公開日 | 2020-07-15 | | 最終更新日 | 2021-01-27 | | 実験手法 | X-RAY DIFFRACTION (3.4011 Å) | | 主引用文献 | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
7QVB
 
 | |
8CWF
 
 | | 200us Temperature-Jump (Light) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | 登録日 | 2022-05-19 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWC
 
 | | 20ns Temperature-Jump (Light) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | 登録日 | 2022-05-19 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
6PD0
 
 | | Crystal structure of the bacterial cellulose synthase subunit G (BcsG) from Escherichia coli, catalytic domain | | 分子名称: | Cellulose biosynthesis protein BcsG, MAGNESIUM ION, ZINC ION | | 著者 | Anderson, A.C, Brenner, T, Weadge, J.T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | TheEscherichia colicellulose synthase subunit G (BcsG) is a Zn2+-dependent phosphoethanolamine transferase.
J.Biol.Chem., 295, 2020
|
|
6ZTP
 
 | | E. coli 70S-RNAP expressome complex in uncoupled state 6 | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Webster, M.W, Takacs, M, Weixlbaumer, A. | | 登録日 | 2020-07-20 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
7AAZ
 
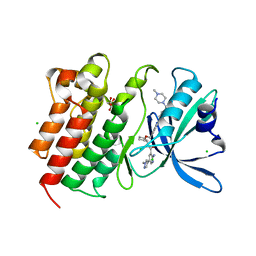 | | Crystal structure of MerTK in complex with a type 1.5 aminopyridine inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-azanyl-~{N}-[(1~{S},2~{S})-2-[[4-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]methoxy]cyclopentyl]-5-(1-methylpyrazol-4-yl)pyridine-3-carboxamide, CHLORIDE ION, ... | | 著者 | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | 登録日 | 2020-09-05 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.855 Å) | | 主引用文献 | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
6PJG
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-97 | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-L-alanyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3 | | 著者 | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | 登録日 | 2019-06-28 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6NB6
 
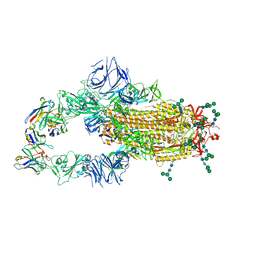 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | 著者 | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2018-12-06 | | 公開日 | 2019-02-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NB7
 
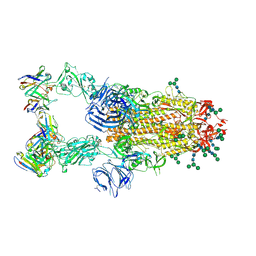 | | SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S230 heavy chain, ... | | 著者 | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2018-12-06 | | 公開日 | 2019-02-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
8FVR
 
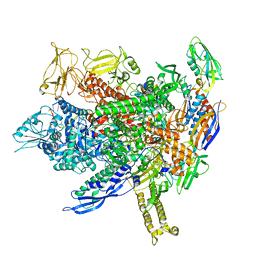 | |
8FVW
 
 | |
