6P58
 
 | | Dark and Steady State-Illuminated Crystal Structure of Cyanobacteriochrome Receptor PixJ at 150K | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | 著者 | Clinger, J.A, Miller, M.D, Buirgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | 登録日 | 2019-05-29 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZTP
 
 | | E. coli 70S-RNAP expressome complex in uncoupled state 6 | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Webster, M.W, Takacs, M, Weixlbaumer, A. | | 登録日 | 2020-07-20 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6UVO
 
 | | Structure of antibody 3G12 bound to the central conserved domain of RSV G | | 分子名称: | 3G12 Fab Heavy chain, 3G12 Fab light chain, Major surface glycoprotein G | | 著者 | Fedechkin, S.O, George, N.L, Nunez Castrejon, A.M, Dillen, J, Kauvar, L.M, DuBois, R.M. | | 登録日 | 2019-11-03 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Conformational Flexibility in Respiratory Syncytial Virus G Neutralizing Epitopes.
J.Virol., 94, 2020
|
|
4LH2
 
 | |
3L1R
 
 | |
5JSX
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+ and uridine-diphosphate-glucose (UDP-Glc) | | 分子名称: | GLYCEROL, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | 著者 | Albesa-Jove, D, Sancho-Vaello, E, Rodrigo-Unzueta, A, Comino, N, Carreras-Gonzalez, A, Arrasate, P, Urresti, S, Guerin, M.E. | | 登録日 | 2016-05-09 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Structural Snapshots and Loop Dynamics along the Catalytic Cycle of Glycosyltransferase GpgS.
Structure, 25, 2017
|
|
6S9U
 
 | | Crystal structure of sucrose 6F-phosphate phosphorylase from Ilumatobacter coccineus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | 著者 | Capra, N, Franceus, J, Desmet, T, Thunnissen, A.M.W.H. | | 登録日 | 2019-07-15 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Comparison of a Promiscuous and a Highly Specific Sucrose 6 F -Phosphate Phosphorylase.
Int J Mol Sci, 20, 2019
|
|
7DBP
 
 | | Linker histone defines structure and self-association behaviour of the 177 bp human chromosome | | 分子名称: | DNA (175-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | 著者 | Wang, S, Vogirala, K.V, Soman, A, Liu, Z.B. | | 登録日 | 2020-10-21 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Linker histone defines structure and self-association behaviour of the 177 bp human chromatosome.
Sci Rep, 11, 2021
|
|
5JT4
 
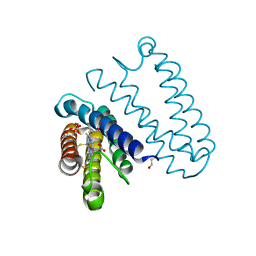 | | L16A mutant of cytochrome c prime from Alcaligenes xylosoxidans: Ferrous state | | 分子名称: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | 著者 | Kekilli, D, Strange, R.W, Hough, M.A. | | 登録日 | 2016-05-09 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
6ZTO
 
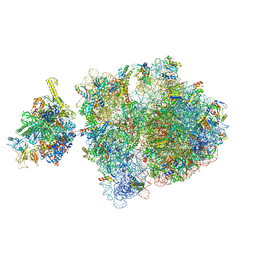 | | E. coli 70S-RNAP expressome complex in uncoupled state 1 | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Webster, M.W, Takacs, M, Weixlbaumer, A. | | 登録日 | 2020-07-20 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6VGV
 
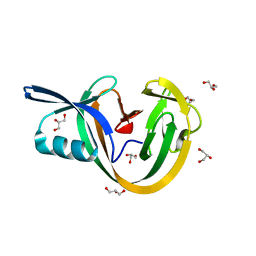 | | Crystal structure of VidaL intein | | 分子名称: | GLYCEROL, VidaL | | 著者 | Burton, A.J, Haugbro, M, Parisi, E, Muir, T.W. | | 登録日 | 2020-01-09 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Live-cell protein engineering with an ultra-short split intein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3B6A
 
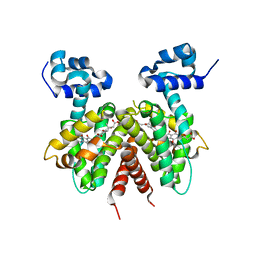 | | Crystal structure of the Streptomyces coelicolor TetR family protein ActR in complex with actinorhodin | | 分子名称: | 2,2'-[(1R,1'R,3S,3'S)-6,6',9,9'-tetrahydroxy-1,1'-dimethyl-5,5',10,10'-tetraoxo-3,3',4,4',5,5',10,10'-octahydro-1H,1'H-8,8'-bibenzo[g]isochromene-3,3'-diyl]diacetic acid, ActR protein | | 著者 | Willems, A.R, Junop, M.S. | | 登録日 | 2007-10-28 | | 公開日 | 2008-02-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA.
J.Mol.Biol., 376, 2008
|
|
6C3R
 
 | |
5JTC
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH)oxygenase and TPR domains in complex with manganese, 2,4-pyridine dicarboxylate and factor X substrate peptide fragment(39mer-4Ser) | | 分子名称: | Aspartyl/asparaginyl beta-hydroxylase, BROMIDE ION, Coagulation factor X, ... | | 著者 | McDonough, M.A, Pfeffer, I. | | 登録日 | 2016-05-09 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Aspartate/asparagine-beta-hydroxylase: a high-throughput mass spectrometric assay for discovery of small molecule inhibitors.
Sci Rep, 10, 2020
|
|
6F9Z
 
 | |
4LJH
 
 | | Crystal Structure of Pseudomonas aeruginosa Lectin LecA Complexed with 1-Methyl-3-indolyl-b-D-galactopyranoside at 1.45 A Resolution | | 分子名称: | 1-methyl-1H-indol-3-ol, CALCIUM ION, PA-I galactophilic lectin, ... | | 著者 | Kadam, R.U, Stocker, A, Reymond, J.L. | | 登録日 | 2013-07-04 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | CH-pi "T-Shape" Interaction with Histidine Explains Binding of Aromatic Galactosides to Pseudomonas aeruginosa Lectin LecA
Acs Chem.Biol., 8, 2013
|
|
8E5U
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 9 | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-(6-{2-nitro-4-[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]anilino}hexyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-08-22 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
4CVO
 
 | | Crystal structure of the N-terminal colied-coil domain of human DNA excision repair protein ERCC-6 | | 分子名称: | DNA EXCISION REPAIR PROTEIN ERCC-6, MAGNESIUM ION | | 著者 | Newman, J.A, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2014-03-28 | | 公開日 | 2014-04-09 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of the N-Terminal Colied-Coil Domain of Human DNA Excision Repair Protein Ercc-6
To be Published
|
|
6BHP
 
 | |
8E3J
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 4 | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{6-[2-nitro-4-(pyrimidin-2-yl)anilino]hexyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-08-17 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8E3P
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 5 | | 分子名称: | (2S,3S,4S,5R)-2-(hydroxymethyl)-1-{6-[3-nitro-5-(pyridin-4-yl)anilino]hexyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-08-17 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
4LKE
 
 | | Crystal Structure of Pseudomonas aeruginosa Lectin LecA Complexed with GalA-WRI at 1.65 A Resolution | | 分子名称: | CALCIUM ION, P-HYDROXYBENZOIC ACID, PA-I galactophilic lectin, ... | | 著者 | Kadam, R.U, Stocker, A, Reymond, J.-L. | | 登録日 | 2013-07-07 | | 公開日 | 2013-12-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.653 Å) | | 主引用文献 | Structure-Based Optimization of the Terminal Tripeptide in Glycopeptide Dendrimer Inhibitors of Pseudomonas aeruginosa Biofilms Targeting LecA.
Chemistry, 19, 2013
|
|
6F7F
 
 | |
7RF8
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2021-07-13 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
6BID
 
 | | 1.15 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | 分子名称: | 3C-like protease, benzyl [(8S,11S,14S)-11-(cyclohexylmethyl)-8-(hydroxymethyl)-5,10,13-trioxo-1,4,9,12,17,18-hexaazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2017-11-01 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
