6VJQ
 
 | |
6VHJ
 
 | |
6VLJ
 
 | |
6VGT
 
 | |
6VE9
 
 | |
6PO6
 
 | | MicroED Structure of a Natural Product VFAThiaGlu | | Descriptor: | YFAThiaGlu | | Authors: | Halaby, S, Gonen, T, Ting, C.P, Funk, M.A, van der Donk, W.A. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Use of a scaffold peptide in the biosynthesis of amino acid-derived natural products.
Science, 365, 2019
|
|
6PQG
 
 | |
6PQF
 
 | |
7JVF
 
 | |
7JU9
 
 | |
2MIJ
 
 | | NMR structure of the S-linked glycopeptide sublancin 168 | | Descriptor: | SPBc2 prophage-derived bacteriocin sublancin-168, beta-D-glucopyranose | | Authors: | Garcia De Gonzalo, C.V, Zhu, L, Oman, T.J, van der Donk, W.A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-03-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the s-linked glycopeptide sublancin 168.
Acs Chem.Biol., 9, 2014
|
|
7RTY
 
 | |
4WD9
 
 | |
4NU5
 
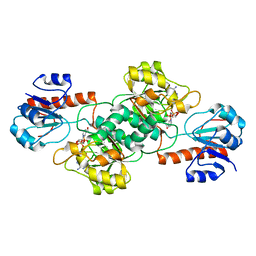 | | Crystal Structure of PTDH R301A | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase | | Authors: | Nair, S.K, Chekan, J.R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
4NU6
 
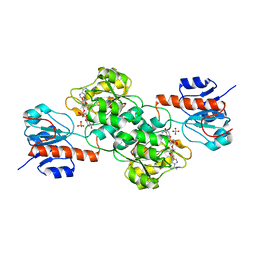 | | Crystal Structure of PTDH R301K | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase, SULFATE ION | | Authors: | Nair, S.K, Chekan, J.R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
4YAR
 
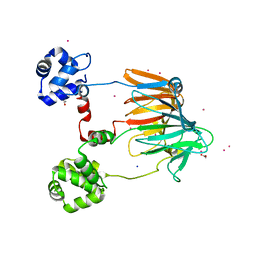 | | 2-Hydroxyethylphosphonate dioxygenase (HEPD) E176H | | Descriptor: | 2-hydroxyethylphosphonate dioxygenase, ACETATE ION, CADMIUM ION, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2015-02-17 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Common Late-Stage Intermediate in Catalysis by 2-Hydroxyethyl-phosphonate Dioxygenase and Methylphosphonate Synthase.
J.Am.Chem.Soc., 137, 2015
|
|
8CZK
 
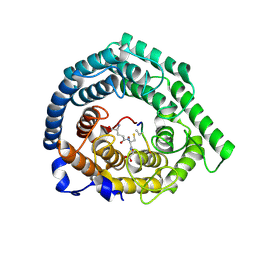 | |
8CZL
 
 | | Human LanCL1 bound to methyl glutathione (MeGSH) | | Descriptor: | Glutathione S-transferase LANCL1, L-GAMMA-GLUTAMYL-S-METHYLCYSTEINYLGLYCINE, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D19
 
 | | Human LanCL1 bound to GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D0V
 
 | | Human LanCL1 C264A mutant bound to GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | Authors: | Ongpipattanakul, C, Nair, S.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8CWX
 
 | |
8HCI
 
 | |
8HI8
 
 | |
8HI7
 
 | |
6WQ1
 
 | | Eukaryotic LanCL2 protein | | Descriptor: | LanC-like protein 2, ZINC ION | | Authors: | Nair, S.K, Garg, N. | | Deposit date: | 2020-04-28 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | LanCLs add glutathione to dehydroamino acids generated at phosphorylated sites in the proteome.
Cell, 184, 2021
|
|
