7XPN
 
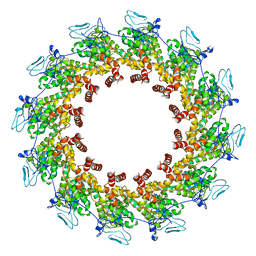 | |
8IXW
 
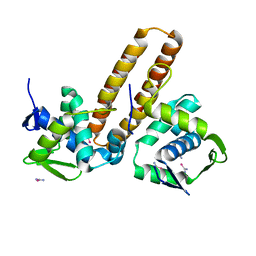 | |
7C5Z
 
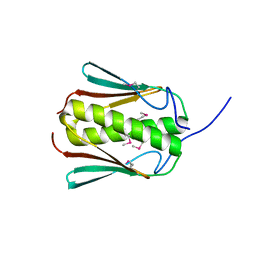 | |
8HCM
 
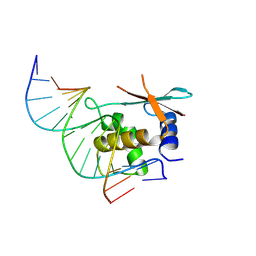 | | zebrafish IRF-11 DBD complex with DNA | | Descriptor: | DNA (5'-D(P*GP*CP*TP*TP*TP*CP*AP*CP*TP*TP*TP*CP*TP*A)-3'), DNA (5'-D(P*TP*AP*GP*AP*AP*AP*GP*TP*GP*AP*AP*AP*GP*C)-3'), Interferon regulatory factor | | Authors: | Wang, Z.X, Zhang, Y.A, Ouyang, S.Y. | | Deposit date: | 2022-11-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of DNA-bound Fish IRF10 and IRF11 Reveal the Determinants of IFN Regulation.
J Immunol., 213, 2024
|
|
8HCL
 
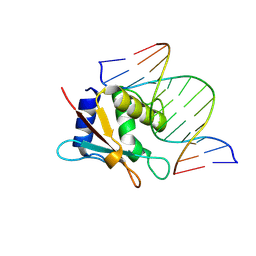 | | zebrafish IRF-10 DBD complex with DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*TP*TP*CP*AP*CP*TP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*AP*GP*TP*GP*AP*AP*AP*GP*T)-3'), Interferon regulatory factor 10 | | Authors: | Wang, Z.X, Zhang, Y.A, Ouyang, S.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structures of DNA-bound Fish IRF10 and IRF11 Reveal the Determinants of IFN Regulation.
J Immunol., 213, 2024
|
|
8HCS
 
 | | zebrafish IRF-11 DBD | | Descriptor: | Interferon regulatory factor | | Authors: | Wang, Z.X, Zhang, Y.A, Ouyang, S.Y. | | Deposit date: | 2022-11-03 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of DNA-bound Fish IRF10 and IRF11 Reveal the Determinants of IFN Regulation.
J Immunol., 213, 2024
|
|
7YG7
 
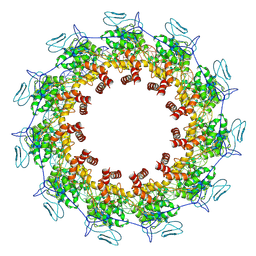 | | Structure of the Spring Viraemia of Carp Virus ribonucleoprotein Complex | | Descriptor: | Nucleoprotein, RNA (99-mer) | | Authors: | Liu, B, Wang, Z.X, Yang, T, Yu, D.Q, Ouyang, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Spring Viraemia of Carp Virus Ribonucleoprotein Complex Reveals Its Assembly Mechanism and Application in Antiviral Drug Screening.
J.Virol., 97, 2023
|
|
7F70
 
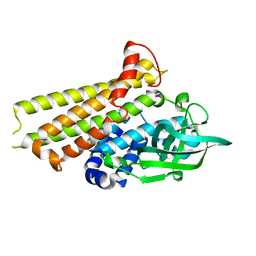 | | Crystal structure of Rv3094c | | Descriptor: | Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y, Zhang, H.T. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
7F74
 
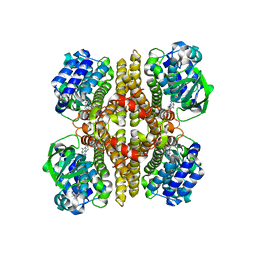 | | Rv3094c in complex with FMN. | | Descriptor: | FLAVIN MONONUCLEOTIDE, Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y. | | Deposit date: | 2021-06-27 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
7F72
 
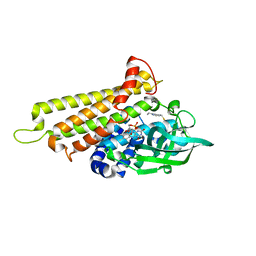 | | Rv3094c in complex with FAD and ETH. | | Descriptor: | 2-ethylpyridine-4-carboximidothioic acid, FLAVIN MONONUCLEOTIDE, Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y. | | Deposit date: | 2021-06-27 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
6WZU
 
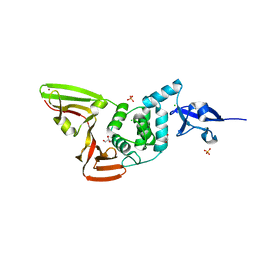 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , P3221 space group | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-14 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
6WRH
 
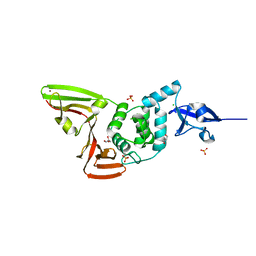 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Welk, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
6XG3
 
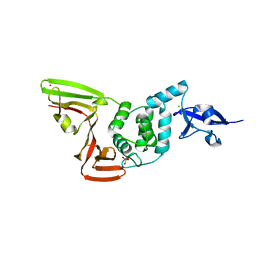 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, at room temperature | | Descriptor: | CHLORIDE ION, Non-structural protein 3, PHOSPHATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIR
 
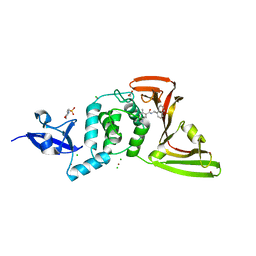 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder457 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIW
 
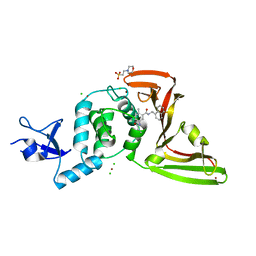 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIV
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIT
 
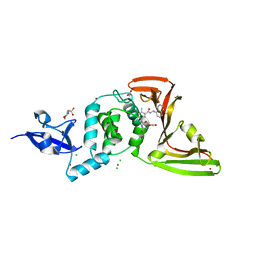 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder495 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(carbamoylcarbamoyl)amino]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
