7ESV
 
 | |
7ESQ
 
 | |
7ESP
 
 | |
7ESO
 
 | |
5W93
 
 | | p130Cas complex with paxillin LD1 | | Descriptor: | Breast cancer anti-estrogen resistance protein 1, Paxillin | | Authors: | Miller, D.J, Zheng, J.J. | | Deposit date: | 2017-06-22 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and functional insights into the interaction between the Cas family scaffolding protein p130Cas and the focal adhesion-associated protein paxillin.
J. Biol. Chem., 292, 2017
|
|
7Y7M
 
 | |
7YMS
 
 | |
7YRH
 
 | |
7YRF
 
 | |
7YV7
 
 | |
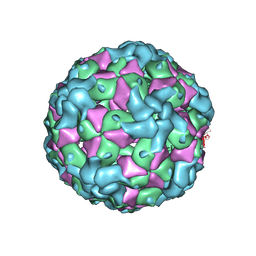
7YV2
 
 | |
4LGH
 
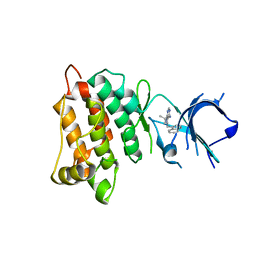 | |
4LGG
 
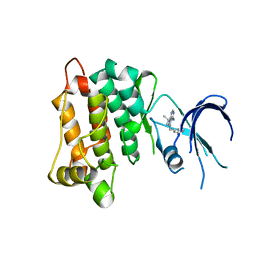 | | Structure of 3MB-PP1 bound to analog-sensitive Src kinase | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Lopez, M.S, Dar, A.C, Shokat, K.M. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-guided inhibitor design expands the scope of analog-sensitive kinase technology.
Acs Chem.Biol., 8, 2013
|
|
8JJC
 
 | | Tubulin-Y62 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(6,7-dimethoxy-3,4-dihydro-1~{H}-isoquinolin-2-yl)-6-(3-methoxyphenyl)pyrimidin-2-amine, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
8JJB
 
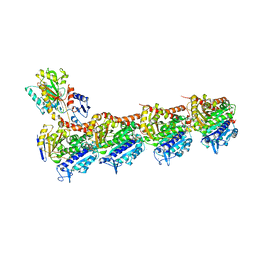 | | Crystal structure of T2R-TTL-Y61 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
6S8N
 
 | | Cryo-EM structure of LptB2FGC in complex with lipopolysaccharide | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, Inner membrane protein yjgQ, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8H
 
 | | Cryo-EM structure of LptB2FG in complex with LPS | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8G
 
 | | Cryo-EM structure of LptB2FGC in complex with AMP-PNP | | Descriptor: | Inner membrane protein yjgQ, LPS export ABC transporter permease LptF, Lipopolysaccharide ABC transporter, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6ZY3
 
 | | Cryo-EM structure of MlaFEDB in complex with phospholipid | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY2
 
 | | Cryo-EM structure of apo MlaFEDB | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, Toluene tolerance protein Ttg2A, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY4
 
 | | Cryo-EM structure of MlaFEDB in complex with ADP | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY9
 
 | | Cryo-EM structure of MlaFEDB in complex with AMP-PNP | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, MAGNESIUM ION, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8JP1
 
 | | Crystal structure of AKR1C3 in complex with DFV | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | Deposit date: | 2023-06-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
8JP2
 
 | | Crystal structure of AKR1C1 in complex with DFV | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | Deposit date: | 2023-06-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
6X9I
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
