2N8X
 
 | | Solution structure of LptE from Pseudomonas Aerigunosa | | Descriptor: | LPS-assembly lipoprotein LptE | | Authors: | Moehle, K, Kocherla, H, Jurt, S, Robinson, J, Zerbe, O, Zerbe, K, Bacsa, B. | | Deposit date: | 2015-10-28 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of LptE from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
6GD5
 
 | | The solution structure of the LptA-Thanatin complex | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin | | Authors: | Moehle, K, Zerbe, O. | | Deposit date: | 2018-04-22 | | Release date: | 2018-11-28 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport inEscherichia coli.
Sci Adv, 4, 2018
|
|
2AHT
 
 | |
1HD6
 
 | | PHEROMONE ER-22, NMR | | Descriptor: | PHEROMONE ER-22 | | Authors: | Luginbuhl, P, Liu, A, Zerbe, O, Ortenzi, C, Luporini, P, Wuthrich, K. | | Deposit date: | 2000-11-09 | | Release date: | 2000-12-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Pheromone Er-22 from Euplotes Raikovi
J.Biomol.NMR, 19, 2001
|
|
1LJV
 
 | | Bovine Pancreatic Polypeptide Bound to DPC Micelles | | Descriptor: | PANCREATIC HORMONE | | Authors: | Lerch, M, Gafner, V, Bader, R, Christen, B, Zerbe, O. | | Deposit date: | 2002-04-22 | | Release date: | 2002-10-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Bovine pancreatic polypeptide (bPP) undergoes significant changes in conformation and dynamics upon binding to DPC micelles.
J.Mol.Biol., 322, 2002
|
|
6GRV
 
 | |
6GW8
 
 | |
6GV7
 
 | |
6GV6
 
 | |
2L61
 
 | | Protein and metal cluster structure of the wheat metallothionein domain g-Ec-1. The second part of the puzzle. | | Descriptor: | CADMIUM ION, EC protein I/II | | Authors: | Loebus, J, Peroza, E.A, Bluethgen, N, Fox, T, Meyer-Klaucke, W, Zerbe, O, Freisinger, E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-05-25 | | Last modified: | 2013-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein and metal cluster structure of the wheat metallothionein domain gamma-E(c)-1: the second part of the puzzle.
J.Biol.Inorg.Chem., 16, 2011
|
|
2MJM
 
 | |
2M0Z
 
 | | cis form of a photoswitchable PDZ domain crosslinked with an azobenzene derivative | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Walser, R, Zerbe, O, Hamm, P. | | Deposit date: | 2012-11-09 | | Release date: | 2013-07-03 | | Last modified: | 2013-08-07 | | Method: | SOLUTION NMR | | Cite: | Kinetic response of a photoperturbed allosteric protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2RLK
 
 | | Refined solution structure of porcine peptide YY (PYY) | | Descriptor: | Peptide YY | | Authors: | Neumoin, A, Mares, J, Lerch-Bader, M, Bader, R, Zerbe, O. | | Deposit date: | 2007-07-21 | | Release date: | 2007-08-14 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Probing the formation of stable tertiary structure in a model miniprotein at atomic resolution: determinants of stability of a helical hairpin
J.Am.Chem.Soc., 129, 2007
|
|
2H3T
 
 | | trans-(4-aminomethyl)phenylazobenzoic acid-aPP bound to DPC micelles | | Descriptor: | (3-{(E)-[3-(AMINOMETHYL)PHENYL]DIAZENYL}PHENYL)ACETIC ACID, Pancreatic hormone | | Authors: | Jurt, S, Aemissegger, A, Guentert, P, Zerbe, O, Hilvert, D. | | Deposit date: | 2006-05-23 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A Photoswitchable Miniprotein Based on the Sequence of Avian Pancreatic Polypeptide
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2H4B
 
 | | cis-4-aminomethylphenylazobenzoic acid-avian pancreatic polypeptide | | Descriptor: | (3-{(Z)-[3-(AMINOMETHYL)PHENYL]DIAZENYL}PHENYL)ACETIC ACID, Pancreatic hormone | | Authors: | Jurt, S, Aemissegger, A, Guentert, P, Zerbe, O, Hilvert, D. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A Photoswitchable Miniprotein Based on the Sequence of Avian Pancreatic Polypeptide
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2H3S
 
 | | cis-Azobenzene-avian pancreatic polypeptide bound to DPC micelles | | Descriptor: | (3-{(Z)-[3-(AMINOMETHYL)PHENYL]DIAZENYL}PHENYL)ACETIC ACID, Pancreatic hormone | | Authors: | Jurt, S, Aemissegger, A, Guentert, P, Zerbe, O, Hilvert, D. | | Deposit date: | 2006-05-23 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A Photoswitchable Miniprotein Based on the Sequence of Avian Pancreatic Polypeptide
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2KAK
 
 | | Solution structure of the beta-E-domain of wheat Ec-1 metallothionein | | Descriptor: | EC protein I/II, ZINC ION | | Authors: | Peroza, E.A, Schmucki, R, Guntert, P, Freisinger, E, Zerbe, O. | | Deposit date: | 2008-11-06 | | Release date: | 2009-05-05 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | The beta(E)-domain of wheat E(c)-1 metallothionein: a metal-binding domain with a distinctive structure.
J.Mol.Biol., 387, 2009
|
|
2K7Z
 
 | |
2K9P
 
 | |
2L62
 
 | | Protein and metal cluster structure of the wheat metallothionein domain g-Ec-1. The second part of the puzzle. | | Descriptor: | EC protein I/II, ZINC ION | | Authors: | Loebus, J, Peroza, E.A, Bluethgen, N, Fox, T, Meyer-Klaucke, W, Zerbe, O, Freisinger, E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-05-25 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Protein and metal cluster structure of the wheat metallothionein domain gamma-E(c)-1: the second part of the puzzle.
J.Biol.Inorg.Chem., 16, 2011
|
|
2L60
 
 | |
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2016-11-09 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2016-11-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
5Y0A
 
 | |
7B6W
 
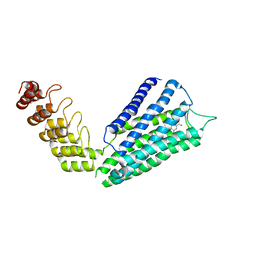 | | Crystal structure of the human alpha1B adrenergic receptor in complex with inverse agonist (+)-cyclazosin | | Descriptor: | Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor, [(4~{a}~{R},8~{a}~{S})-4-(4-azanyl-6,7-dimethoxy-quinazolin-2-yl)-2,3,4~{a},5,6,7,8,8~{a}-octahydroquinoxalin-1-yl]-(furan-2-yl)methanone | | Authors: | Deluigi, M, Morstein, L, Hilge, M, Schuster, M, Merklinger, L, Klipp, A, Scott, D.J, Plueckthun, A. | | Deposit date: | 2020-12-08 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Crystal structure of the alpha 1B -adrenergic receptor reveals molecular determinants of selective ligand recognition.
Nat Commun, 13, 2022
|
|
