6CN2
 
 | | Crystal structure of zebrafish Phosphatidylinositol-4-phosphate 5- kinase alpha isoform D236N with bound ATP/Ca2+ | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Phosphatidylinositol-4-phosphate 5-kinase, ... | | 著者 | Zeng, X, Sui, D, Hu, J. | | 登録日 | 2018-03-07 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Structural insights into lethal contractural syndrome type 3 (LCCS3) caused by a missense mutation of PIP5K gamma.
Biochem. J., 475, 2018
|
|
6CMW
 
 | | Crystal structure of zebrafish Phosphatidylinositol-4-phosphate 5- kinase alpha isoform with bound ATP/Ca2+ | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Phosphatidylinositol-4-phosphate 5-kinase, ... | | 著者 | Zeng, X, Sui, D, Hu, J. | | 登録日 | 2018-03-06 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural insights into lethal contractural syndrome type 3 (LCCS3) caused by a missense mutation of PIP5K gamma.
Biochem. J., 475, 2018
|
|
6CN3
 
 | |
8HJA
 
 | | The crystal structure of syn_CdgR-(c-di-GMP) from Synechocystis sp. PCC 6803 | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), c-di-GMP receptor | | 著者 | Zeng, X, Peng, Y.J. | | 登録日 | 2022-11-22 | | 公開日 | 2023-03-29 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | A c-di-GMP binding effector controls cell size in a cyanobacterium.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4NC4
 
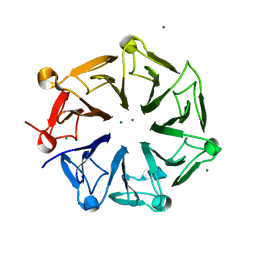 | | Crystal structure of photoreceptor AtUVR8 mutant W285F and light-induced structural changes at 120K | | 分子名称: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | 著者 | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | 登録日 | 2013-10-23 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NBM
 
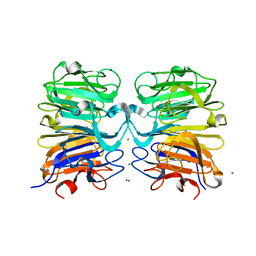 | | Crystal structure of UVB photoreceptor UVR8 and light-induced structural changes at 180K | | 分子名称: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | 著者 | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | 登録日 | 2013-10-23 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NAA
 
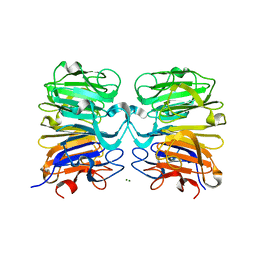 | | Crystal structure of UVB photoreceptor UVR8 from Arabidopsis thaliana and UV-induced structural changes at 120K | | 分子名称: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | 著者 | Yang, X, Zeng, X, Ren, Z, Zhao, K.H. | | 登録日 | 2013-10-22 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
8BLT
 
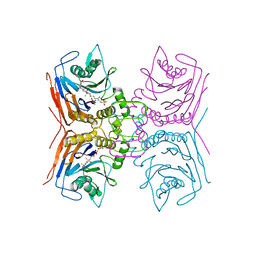 | | Structure of Lactobacillus salivarius (Ls) bile salt hydrolase(BSH) in complex with taurocholate (TCA) | | 分子名称: | Bile salt hydrolase, TAUROCHOLIC ACID | | 著者 | Karlov, D.S, Long, S.L, Zeng, X, Xu, F, Lal, K, Cao, L, Hayoun, K, Lin, J, Joyce, S.A, Tikhonova, I.G. | | 登録日 | 2022-11-10 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Characterization of the mechanism of bile salt hydrolase substrate specificity by experimental and computational analyses.
Structure, 31, 2023
|
|
8BLS
 
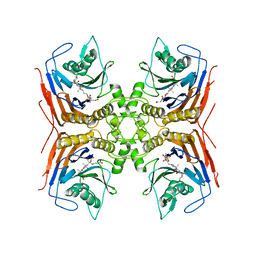 | | Structure of Lactobacillus salivarius (Ls) bile salt hydrolase(BSH) in complex with Glycocholate (GCA) | | 分子名称: | Bile salt hydrolase, GLYCOCHOLIC ACID | | 著者 | Karlov, D.S, Long, S.L, Zeng, X, Xu, F, Lal, K, Cao, L, Hayoun, K, Lin, J, Joyce, S.A, Tikhonova, I.G. | | 登録日 | 2022-11-10 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Characterization of the mechanism of bile salt hydrolase substrate specificity by experimental and computational analyses.
Structure, 31, 2023
|
|
4GBY
 
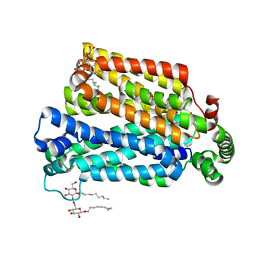 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose | | 分子名称: | D-xylose-proton symporter, beta-D-xylopyranose, nonyl beta-D-glucopyranoside | | 著者 | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | 登録日 | 2012-07-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.808 Å) | | 主引用文献 | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GC0
 
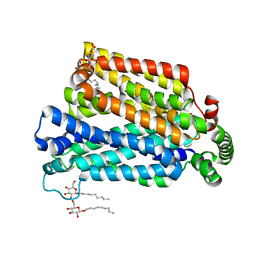 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | 分子名称: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | 著者 | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | 登録日 | 2012-07-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GBZ
 
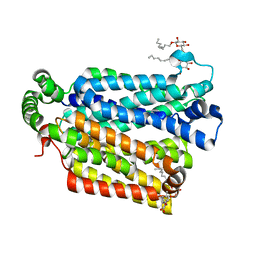 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose | | 分子名称: | D-xylose-proton symporter, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | 著者 | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | 登録日 | 2012-07-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.894 Å) | | 主引用文献 | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
7F77
 
 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans | | 分子名称: | Glutamate dehydrogenase | | 著者 | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | 登録日 | 2021-06-28 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.086 Å) | | 主引用文献 | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
7F79
 
 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans in complex with alpha-ketoglutarate and NADPH | | 分子名称: | 2-OXOGLUTARIC ACID, GLYCEROL, Glutamate dehydrogenase, ... | | 著者 | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | 登録日 | 2021-06-28 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
7VC8
 
 | | Complex structure of AtHPPD with inhibitor PYQ3 | | 分子名称: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, pyren-1-yl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate | | 著者 | Yang, G.F, Lin, H.Y. | | 登録日 | 2021-09-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.606 Å) | | 主引用文献 | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7WJ8
 
 | | Complex structure of AtHPPD-PyQ1 | | 分子名称: | 2-pyren-1-yloxyethyl 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]ethanoate, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | 著者 | Yang, G.-F, Lin, H.-Y, Dong, J. | | 登録日 | 2022-01-05 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.805 Å) | | 主引用文献 | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7WJJ
 
 | | Complex structure of AtHPPD-PyQ2 | | 分子名称: | 2-[1,5-dimethyl-2,4-bis(oxidanylidene)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazolin-3-yl]-N-(2-pyren-1-yloxyethyl)ethanamide, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | 著者 | Yang, G.-F, Lin, H.-Y, Dong, J. | | 登録日 | 2022-01-06 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.603 Å) | | 主引用文献 | Design of an HPPD fluorescent probe and visualization of plant responses to abiotic stress
Adv Agrochem, 2022
|
|
7VSQ
 
 | |
7VSP
 
 | |
7DNR
 
 | |
3L1L
 
 | |
5KCM
 
 | | Crystal structure of iron-sulfur cluster containing photolyase PhrB mutant I51W | | 分子名称: | (6-4) photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Yang, X, Bowatte, K, Zhang, F, Lamparter, T. | | 登録日 | 2016-06-06 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.149 Å) | | 主引用文献 | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
6DD6
 
 | |
5LFA
 
 | | Crystal structure of iron-sulfur cluster containing bacterial (6-4) photolyase PhrB - Y424F mutant with impaired DNA repair activity | | 分子名称: | (6-4) photolyase, 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Kwiatkowski, D, Zhang, F, Krauss, N, Lamparter, T, Scheerer, P. | | 登録日 | 2016-06-30 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
4ZYL
 
 | |
