6LON
 
 | | Crystal structure of HPSG | | Descriptor: | (2~{S})-2,3-bis(oxidanyl)propane-1-sulfonic acid, GLYCEROL, PFL2/glycerol dehydratase family glycyl radical enzyme, ... | | Authors: | Liu, J, Zhang, Y, Yuchi, Z. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two radical-dependent mechanisms for anaerobic degradation of the globally abundant organosulfur compound dihydroxypropanesulfonate.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LT9
 
 | |
6M2W
 
 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
6LQK
 
 | | Crystal structure of honeybee RyR NTD | | Descriptor: | MAGNESIUM ION, ryanodine receptor | | Authors: | Zhou, Y, Lin, L, Yuchi, Z. | | Deposit date: | 2020-01-13 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Crystal structure of the N-terminal domain of ryanodine receptor from the honeybee, Apis mellifera.
Insect Biochem.Mol.Biol., 125, 2020
|
|
7E7L
 
 | | The crystal structure of arylacetate decarboxylase from Olsenella scatoligenes. | | Descriptor: | 4-HYDROXYPHENYLACETATE, Hydroxyphenylacetic acid decarboxylase | | Authors: | Lu, Q, Duan, Y, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The Glycyl Radical Enzyme Arylacetate Decarboxylase from Olsenella scatoligenes
Acs Catalysis, 11, 2021
|
|
7EEV
 
 | | Structure of UTP cyclohydrolase | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, GTP cyclohydrolase II, ZINC ION | | Authors: | Zhang, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-07-28 | | Last modified: | 2022-09-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical investigation of UTP cyclohydrolase
Acs Catalysis, 2021
|
|
7EJ3
 
 | | UTP cyclohydrolase | | Descriptor: | DIPHOSPHATE, GLYCINE, GTP cyclohydrolase II, ... | | Authors: | Zhang, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-04-01 | | Release date: | 2022-09-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical investigation of UTP cyclohydrolase
Acs Catalysis, 2021
|
|
7VUA
 
 | | Anaerobic hydroxyproline degradation involving C-N cleavage by a glycyl radical enzyme | | Descriptor: | (4S)-4-hydroxy-D-proline, HplG | | Authors: | Duan, Y, Lu, Q, Yuchi, Z, Zhang, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Anaerobic Hydroxyproline Degradation Involving C-N Cleavage by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 144, 2022
|
|
7F2B
 
 | |
7F2E
 
 | | SARS-CoV-2 nucleocapsid protein C-terminal domain (dodecamer) | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Liu, C, Jiang, H. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the SARS-CoV-2 nucleocapsid protein C-terminal domain and development of nucleocapsid-targeting nanobodies.
Febs J., 289, 2022
|
|
5ZXL
 
 | | Structure of GldA from E.coli | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycerol dehydrogenase, ... | | Authors: | Zhang, J, Lin, L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structure of glycerol dehydrogenase (GldA) from Escherichia coli.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
6MM7
 
 | |
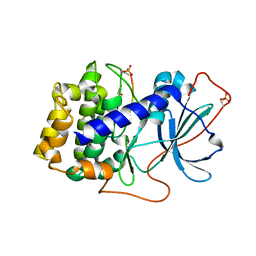
6MM5
 
 | |
6MM6
 
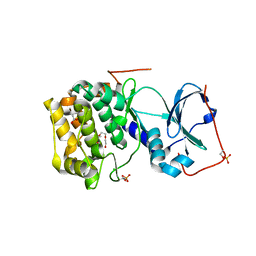 | |
6MM8
 
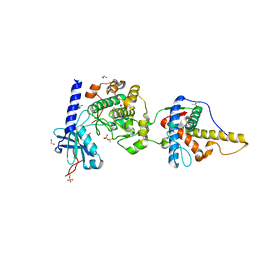 | |
7X05
 
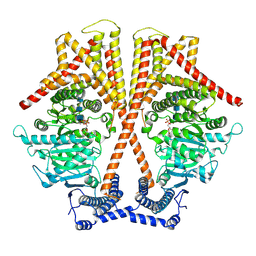 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with the nascent chitooligosaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin synthase, MANGANESE (II) ION, ... | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7WJN
 
 | | CryoEM structure of chitin synthase 1 mutant E495A from Phytophthora sojae complexed with UDP-GlcNAc | | Descriptor: | Chitin synthase, MANGANESE (II) ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7WJM
 
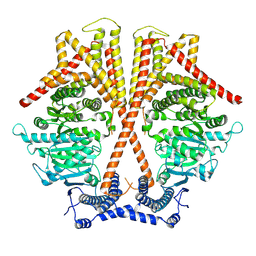 | |
7WJO
 
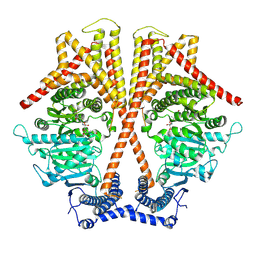 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7X06
 
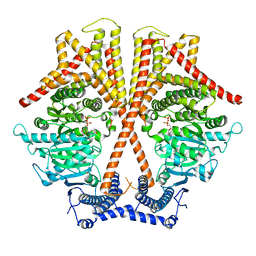 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with UDP | | Descriptor: | Chitin synthase, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
