8VQ1
 
 | | Pseudomonas fluorescens G150T isocyanide hydratase at 298 K XFEL data, thioimidate intermediate | | Descriptor: | CHLORIDE ION, Isonitrile hydratase InhA, N-(4-nitrophenyl)methanimine | | Authors: | Wilson, M.A, Smith, N, Dasgupta, M, Dolamore, C. | | Deposit date: | 2024-01-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in an enzyme ensemble during catalysis observed by high-resolution XFEL crystallography.
Sci Adv, 10, 2024
|
|
6Y0P
 
 | | isopenicillin N synthase in complex with IPN and Fe using FT-SSX methods | | Descriptor: | FE (III) ION, ISOPENICILLIN N, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
7QX4
 
 | | mosquitocidal Cry11Aa determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX6
 
 | | mosquitocidal Cry11Aa-E583Q determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX5
 
 | | mosquitocidal Cry11Aa-Y449F determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX7
 
 | | mosquitocidal Cry11Aa-F17Y determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
8FG7
 
 | |
8FRC
 
 | |
8FRA
 
 | | Mouse acidic mammalian chitinase, catalytic domain in complex with diacetylchitobiose at pH 5.60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acidic mammalian chitinase, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-01-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
8FRB
 
 | | Mouse acidic mammalian chitinase, catalytic domain in complex with N,N'-diacetylchitobiose at pH 5.25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetylamino-2-deoxy-alpha-L-idopyranose, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-01-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
8FRD
 
 | |
8FR9
 
 | |
8FRG
 
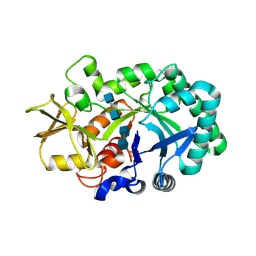 | |
8GCA
 
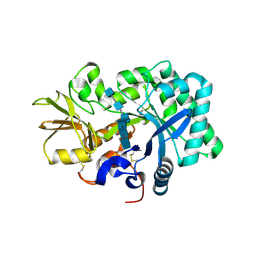 | | Mouse acidic mammalian chitinase, catalytic domain in complex with N,N',N''-triacetylchitotriose at pH 4.74 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acidic mammalian chitinase, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
7QYD
 
 | | mosquitocidal Cry11Ba determined at pH 6.5 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Ba | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7R1E
 
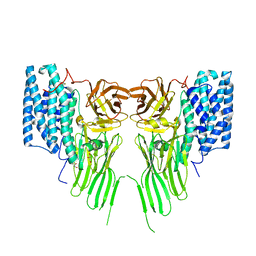 | | Mosquitocidal Cry11Ba determined at pH 10.4 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | GLYCEROL, Pesticidal crystal protein Cry11Ba | | Authors: | Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7LVK
 
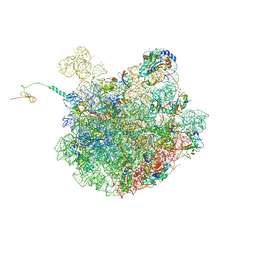 | | Cfr-modified 50S subunit from Escherichia coli | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Stojkovic, V, Myasnikov, A.G, Frost, A, Fujimori, D.G. | | Deposit date: | 2021-02-25 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Investigating antibiotic resistance of a ribosomal-RNA methylating enzyme through directed evolution
To Be Published
|
|
7M75
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M78
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M76
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
5S20
 
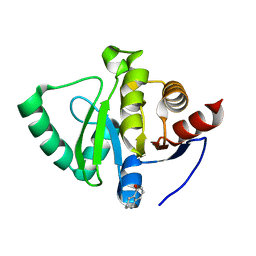 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with PB1827975385 | | Descriptor: | (5R)-5-amino-5,6,7,8-tetrahydronaphthalen-1-ol, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.037 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2K
 
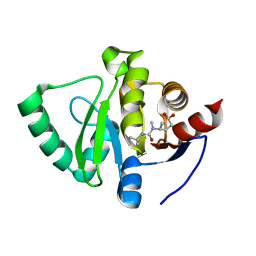 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z445856640 | | Descriptor: | N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]-N-methyl-N'-propan-2-ylurea, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S30
 
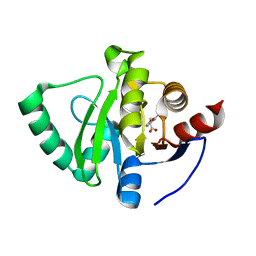 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z65532537 | | Descriptor: | (2R)-2-(2-fluorophenoxy)propanoic acid, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3K
 
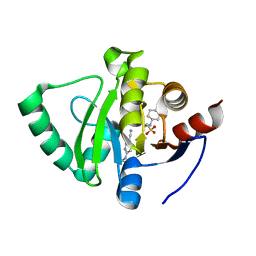 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z219104216 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 6-(ethylamino)pyridine-3-carbonitrile, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S46
 
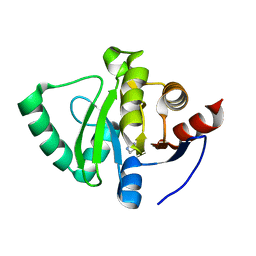 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z57131035 | | Descriptor: | Non-structural protein 3, imidazolidine-2,4-dione | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.191 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
