1UK4
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) Complexed With An Inhibitor | | 分子名称: | 3C-like proteinase nsp5, 5-mer peptide of inhibitor | | 著者 | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | 登録日 | 2003-08-14 | | 公開日 | 2003-11-18 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7D60
 
 | | Cryo-EM Structure of human CALHM5 in the presence of rubidium red | | 分子名称: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | 著者 | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-23 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7D65
 
 | | Cryo-EM Structure of human CALHM5 in the presence of Ca2+ | | 分子名称: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | 著者 | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | 登録日 | 2020-09-29 | | 公開日 | 2020-12-23 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7D61
 
 | | Cryo-EM Structure of human CALHM5 in the presence of EDTA | | 分子名称: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | 著者 | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-23 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
1ZXX
 
 | | The crystal structure of phosphofructokinase from Lactobacillus delbrueckii | | 分子名称: | 6-phosphofructokinase, SULFATE ION | | 著者 | Paricharttanakul, N.M, Ye, S, Menefee, A.L, Javid-Majd, F, Sacchettini, J.C, Reinhart, G.D. | | 登録日 | 2005-06-09 | | 公開日 | 2005-11-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Kinetic and Structural Characterization of Phosphofructokinase from Lactobacillus bulgaricus.
Biochemistry, 44, 2005
|
|
2AHY
 
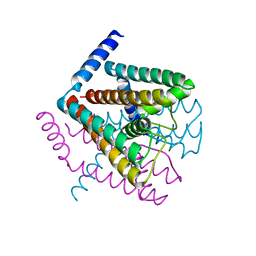 | | Na+ complex of the NaK Channel | | 分子名称: | CALCIUM ION, Potassium channel protein, SODIUM ION | | 著者 | Shi, N, Ye, S, Alam, A, Chen, L, Jiang, Y. | | 登録日 | 2005-07-28 | | 公開日 | 2006-02-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Atomic structure of a Na+- and K+-conducting channel.
Nature, 440, 2006
|
|
2AIU
 
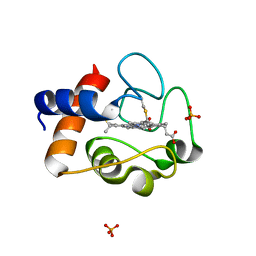 | | Crystal Structure of Mouse Testicular Cytochrome C at 1.6 Angstrom | | 分子名称: | Cytochrome c, testis-specific, PHOSPHATE ION, ... | | 著者 | Liu, Z, Ye, S, Lin, H, Rao, Z, Liu, X.J. | | 登録日 | 2005-08-01 | | 公開日 | 2006-07-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Remarkably high activities of testicular cytochrome c in destroying reactive oxygen species and in triggering apoptosis
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AHZ
 
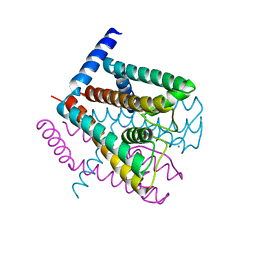 | | K+ complex of the NaK Channel | | 分子名称: | CALCIUM ION, POTASSIUM ION, Potassium channel protein | | 著者 | Shi, N, Ye, S, Alam, A, Chen, L, Jiang, Y. | | 登録日 | 2005-07-28 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Atomic structure of a Na+- and K+-conducting channel.
Nature, 440, 2006
|
|
5B63
 
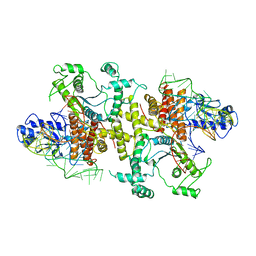 | | Crystal structures of E.coli arginyl-tRNA synthetase (ArgRS) in complex with substrate tRNA(Arg) | | 分子名称: | Arginine--tRNA ligase, tRNA-Arg | | 著者 | Zhou, M, Ye, S, Stephen, P, Zhang, R, Wang, E.D, Giege, R, Lin, S.X. | | 登録日 | 2016-05-24 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of E.coli arginyl-tRNA synthetase (ArgRS) in complex with substrate tRNA(Arg)
To Be Published
|
|
5COA
 
 | | Crystal structure of iridoid synthase at 2.2-angstrom resolution | | 分子名称: | HEXAETHYLENE GLYCOL, Iridoid synthase, SULFATE ION | | 著者 | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | 登録日 | 2015-07-20 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
5COB
 
 | | Crystal structure of iridoid synthase in complex with NADP+ and 8-oxogeranial at 2.65-angstrom resolution | | 分子名称: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, Iridoid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | 登録日 | 2015-07-20 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
4F7G
 
 | |
4F7H
 
 | | The crystal structure of kindlin-2 pleckstrin homology domain in free form | | 分子名称: | Fermitin family homolog 2, S,R MESO-TARTARIC ACID | | 著者 | Liu, Y, Zhu, Y, Qin, J, Ye, S, Zhang, R. | | 登録日 | 2012-05-16 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of kindlin-2 PH domain reveals a conformational transition for its membrane anchoring and regulation of integrin activation.
Protein Cell, 3, 2012
|
|
4FNV
 
 | | Crystal Structure of Heparinase III | | 分子名称: | Heparinase III protein, heparitin sulfate lyase | | 著者 | Dong, W, Ye, S. | | 登録日 | 2012-06-20 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis of heparan sulfate-specific degradation by heparinase III.
Protein Cell, 3, 2012
|
|
4GX2
 
 | | GsuK channel bound to NAD | | 分子名称: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, ... | | 著者 | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | 登録日 | 2012-09-03 | | 公開日 | 2012-12-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
4GX1
 
 | | Crystal structure of the GsuK bound to ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, PHOSPHATE ION, ... | | 著者 | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | 登録日 | 2012-09-03 | | 公開日 | 2012-12-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
4H5Y
 
 | | High-resolution crystal structure of Legionella pneumophila LidA (60-594) | | 分子名称: | LidA protein, substrate of the Dot/Icm system | | 著者 | An, X, Ye, S, Liu, Y, Zheng, X, Zhang, R. | | 登録日 | 2012-09-19 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of LidA, a translocated substrate of the Legionella pneumophila type IV secretion system.
Protein Cell, 4, 2013
|
|
4GX0
 
 | | Crystal structure of the GsuK L97D mutant | | 分子名称: | CALCIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | 登録日 | 2012-09-03 | | 公開日 | 2012-12-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
4GX5
 
 | | GsuK Channel | | 分子名称: | CALCIUM ION, POTASSIUM ION, TrkA domain protein, ... | | 著者 | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | 登録日 | 2012-09-03 | | 公開日 | 2012-12-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
8X84
 
 | |
8X83
 
 | |
8X82
 
 | |
7WKS
 
 | | Apo state of AtPIN3 | | 分子名称: | Auxin efflux carrier component 3 | | 著者 | Su, N. | | 登録日 | 2022-01-11 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7WKW
 
 | | NPA bound state of AtPIN3 | | 分子名称: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 3 | | 著者 | Su, N. | | 登録日 | 2022-01-11 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7XXB
 
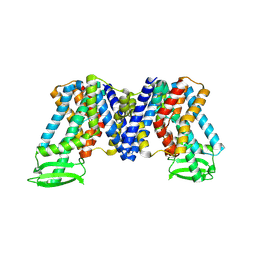 | | IAA bound state of AtPIN3 | | 分子名称: | 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 3 | | 著者 | Su, N. | | 登録日 | 2022-05-29 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
