1V54
 
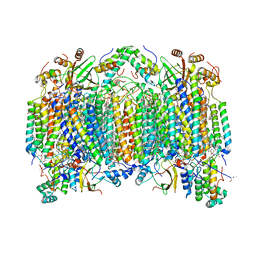 | | Bovine heart cytochrome c oxidase at the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1V4N
 
 | | Structure of 5'-deoxy-5'-methylthioadenosine phosphorylase homologue from Sulfolobus tokodaii | | Descriptor: | 271aa long hypothetical 5'-methylthioadenosine phosphorylase | | Authors: | Kitago, Y, Yasutake, Y, Sakai, N, Tsujimura, M, Yao, M, Watanabe, N, Kawarabayasi, Y, Tanaka, I. | | Deposit date: | 2003-11-14 | | Release date: | 2005-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Sulfolobus tokodaii MTAP
To be Published
|
|
1VBF
 
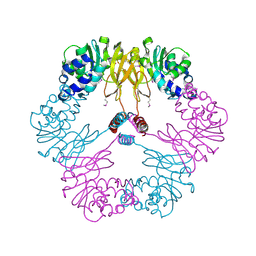 | | Crystal structure of protein L-isoaspartate O-methyltransferase homologue from Sulfolobus tokodaii | | Descriptor: | 231aa long hypothetical protein-L-isoaspartate O-methyltransferase | | Authors: | Tanaka, Y, Tsumoto, K, Yasutake, Y, Umetsu, M, Yao, M, Tanaka, I, Fukada, H, Kumagai, I. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How Oligomerization Contributes to the Thermostability of an Archaeon Protein: PROTEIN L-ISOASPARTYL-O-METHYLTRANSFERASE FROM SULFOLOBUS TOKODAII
J.Biol.Chem., 279, 2004
|
|
1WMI
 
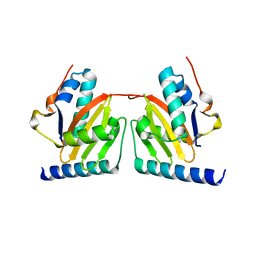 | | Crystal structure of archaeal RelE-RelB complex from Pyrococcus horikoshii OT3 | | Descriptor: | hypothetical protein PHS013, hypothetical protein PHS014 | | Authors: | Takagi, H, Kakuta, Y, Kamachi, R, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2004-07-09 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WNU
 
 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with L-serine | | Descriptor: | SERINE, ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2004-08-09 | | Release date: | 2005-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1X0T
 
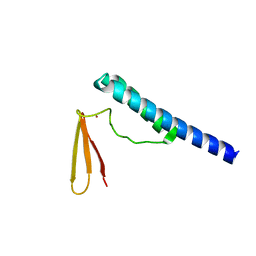 | | Crystal structure of ribonuclease P protein Ph1601p from Pyrococcus horikoshii OT3 | | Descriptor: | Ribonuclease P protein component 4, ZINC ION | | Authors: | Kakuta, Y, Ishimatsu, I, Numata, T, Kimura, K, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2005-03-29 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Ribonuclease P Protein Ph1601p from Pyrococcus horikoshii OT3: An Archaeal Homologue of Human Nuclear Ribonuclease P Protein Rpp21(,)
Biochemistry, 44, 2005
|
|
1WXO
 
 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with zinc | | Descriptor: | ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WZZ
 
 | | Structure of endo-beta-1,4-glucanase CMCax from Acetobacter xylinum | | Descriptor: | Probable endoglucanase, SULFATE ION | | Authors: | Yasutake, Y, Kawano, S, Tajima, K, Yao, M, Satoh, Y, Munekata, M, Tanaka, I, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-10 | | Release date: | 2006-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of the Acetobacter xylinum endo-beta-1,4-glucanase CMCax required for cellulose biosynthesis.
Proteins, 64, 2006
|
|
8IN3
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN4
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai | | Descriptor: | 25 kDa polyphenol-binding protein, ACETYL GROUP, GLYCEROL | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN6
 
 | | Eisenia hydrolysis-enhancing protein from Aplysia kurodai complex with tannic acid | | Descriptor: | 25 kDa polyphenol-binding protein, BETA-1,2,3,4,6-PENTA-O-GALLOYL-D-GLUCOPYRANOSE | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
8IN1
 
 | | beta-glucosidase protein from Aplysia kurodai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-Glucosidase, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, X.M, Ye, Y.X, Kato, K, Yu, J, Yao, M. | | Deposit date: | 2023-03-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of EHEP-mediated offense against phlorotannin-induced defense from brown algae to protect aku BGL activity.
Elife, 12, 2023
|
|
1UMJ
 
 | | Crystal structure of Pyrococcus horikoshii CutA in the presence of 3M guanidine hydrochloride | | Descriptor: | GUANIDINE, periplasmic divalent cation tolerance protein CutA | | Authors: | Tanaka, Y, Tsumoto, K, Yasutake, Y, Sakai, N, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2003-10-02 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for guanidine-protein side chain interactions: crystal structure of CutA from Pyrococcus horikoshii in 3M guanidine hydrochloride
Biochem.Biophys.Res.Commun., 323, 2004
|
|
8H25
 
 | | Lacticaseibacillus casei GH35 beta-galactosidase LBCZ_0230 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saburi, W, Ota, T, Kato, K, Tagami, T, Yamashita, K, Yao, M, Mori, H. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Function and structure of Lacticaseibacillus cases GH35 beta-galactosidase LBCZ_0230 with high hydrolytic activity to lactose-N-biose I and galactic-N-biose.
J.Appl.Glycosci., 70, 2023
|
|
8IBK
 
 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258G in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
8IDS
 
 | | Crystal structure of Bacillus sp. AHU2216 GH13_31 Alpha-glucosidase E256Q/N258P in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Auiewiriyanukul, W, Saburi, W, Yu, J, Kato, K, Yao, M, Mori, H. | | Deposit date: | 2023-02-14 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alteration of Substrate Specificity and Transglucosylation Activity of GH13_31 alpha-Glucosidase from Bacillus sp. AHU2216 through Site-Directed Mutagenesis of Asn258 on beta → alpha Loop 5.
Molecules, 28, 2023
|
|
1IXL
 
 | | Crystal structure of uncharacterized protein PH1136 from Pyrococcus horikoshii | | Descriptor: | hypothetical protein PH1136 | | Authors: | Tajika, Y, Sakai, N, Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2002-06-27 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of conserved protein PH1136 from Pyrococcus horikoshii.
Proteins, 55, 2004
|
|
1J08
 
 | | Crystal structure of glutaredoxin-like protein from Pyrococcus horikoshii | | Descriptor: | glutaredoxin-like protein | | Authors: | Tanaka, Y, Tanabe, E, Tsumoto, K, Kumagai, I, Yao, M, Tanaka, I. | | Deposit date: | 2002-11-11 | | Release date: | 2003-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein disulfide isomerase from hyperthermophile as an additives of refolding of an immunoglobulin-folded protein
To be Published
|
|
1J31
 
 | | Crystal Structure of Hypothetical Protein PH0642 from Pyrococcus horikoshii | | Descriptor: | ACETATE ION, Hypothetical protein PH0642 | | Authors: | Sakai, N, Tajika, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-01-16 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of hypothetical protein PH0642 from Pyrococcus horikoshii at 1.6A resolution.
Proteins, 57, 2004
|
|
1IXK
 
 | | Crystal Structure Analysis of Methyltransferase Homolog Protein from Pyrococcus Horikoshii | | Descriptor: | Methyltransferase | | Authors: | Ishikawa, I, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2002-06-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human p120 homologue protein PH1374 from Pyrococcus horikoshii
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
1J2V
 
 | | Crystal Structure of CutA1 from Pyrococcus Horikoshii | | Descriptor: | 102AA long hypothetical periplasmic divalent cation tolerance protein CUTA | | Authors: | Tanaka, Y, Sakai, N, Yasutake, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural implications for heavy metal-induced reversible assembly and aggregation of a protein: the case of Pyrococcus horikoshii CutA.
Febs Lett., 556, 2004
|
|
5X3J
 
 | | Kfla1895 D451A mutant in complex with cyclobis-(1->6)-alpha-nigerosyl | | Descriptor: | Cyclic alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, GLYCEROL, Glycoside hydrolase family 31, ... | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycoside hydrolase mutant in complex with substrate
To Be Published
|
|
5X3I
 
 | | Kfla1895 D451A mutant | | Descriptor: | GLYCEROL, Glycoside hydrolase family 31, SULFATE ION | | Authors: | Tanaka, Y, Chen, M, Tagami, T, Yao, M, Kimura, A. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycoside hydrolase mutant
To Be Published
|
|
5X6C
 
 | | Crystal structure of SepRS-SepCysE from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, O-phosphoserine--tRNA(Cys) ligase, SULFATE ION, ... | | Authors: | Chen, M, Kato, K, Yao, M. | | Deposit date: | 2017-02-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Structural basis for tRNA-dependent cysteine biosynthesis
Nat Commun, 8, 2017
|
|
5X6B
 
 | |
