2A7T
 
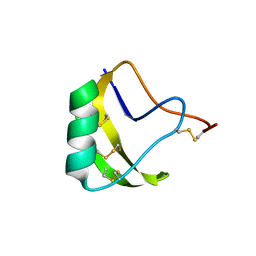 | | Crystal Structure of a novel neurotoxin from Buthus tamalus at 2.2A resolution. | | Descriptor: | Neurotoxin | | Authors: | Ethayathulla, A.S, Sharma, M, Saravanan, K, Sharma, S, Kaur, P, Yadav, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2005-07-06 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a highly acidic neurotoxin from scorpion Buthus tamulus at 2.2A resolution reveals novel structural features.
J.Struct.Biol., 155, 2006
|
|
1RMR
 
 | | Crystal Structure of Schistatin, a Disintegrin Homodimer from saw-scaled Viper (Echis carinatus) at 2.5 A resolution | | Descriptor: | Disintegrin schistatin | | Authors: | Bilgrami, S, Tomar, S, Yadav, S, Kaur, P, Kumar, J, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of schistatin, a disintegrin homodimer from saw-scaled viper (Echis carinatus) at 2.5 A resolution
J.Mol.Biol., 341, 2004
|
|
6LVD
 
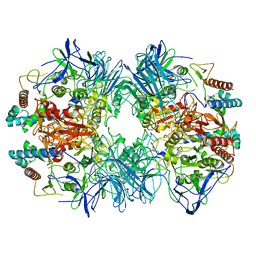 | | Structure of Dimethylformamidase, tetramer, Y440A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1TEJ
 
 | | Crystal structure of a disintegrin heterodimer at 1.9 A resolution. | | Descriptor: | disintegrin chain A, disintegrin chain B | | Authors: | Bilgrami, S, Kaur, P, Yadav, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Disintegrin Heterodimer from Saw-Scaled Viper (Echis carinatus) at 1.9 A Resolution
Biochemistry, 44, 2005
|
|
6LVE
 
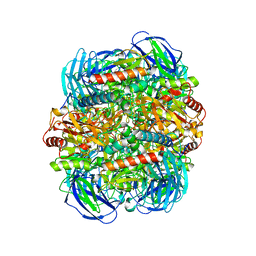 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVC
 
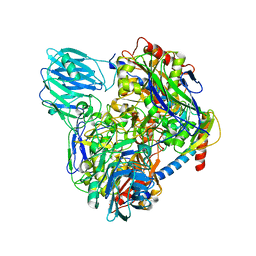 | | Structure of Dimethylformamidase, dimer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVB
 
 | | Structure of Dimethylformamidase, tetramer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1TFM
 
 | | CRYSTAL STRUCTURE OF A RIBOSOME INACTIVATING PROTEIN IN ITS NATURALLY INHIBITED FORM | | Descriptor: | 2-AMINO-4-ISOPROPYL-PTERIDINE-6-CARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mishra, V, Bilgrami, S, Paramasivam, M, Yadav, S, Sharma, R.S, Kaur, P, Srinivasan, A, Babu, C.R, Singh, T.P. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF A RIBOSOME INACTIVATING PROTEIN IN ITS NATURALLY INHIBITED FORM
To be Published
|
|
1TFV
 
 | | CRYSTAL STRUCTURE OF A BUFFALO SIGNALING GLYCOPROTEIN (SPB-40) SECRETED DURING INVOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, mammary gland protein 40 | | Authors: | Bilgrami, S, Saravanan, K, Yadav, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CRYSTAL STRUCTURE OF A BUFFALO SIGNALING GLYCOPROTEIN (SPB-40)
SECRETED DURING INVOLUTION
To be Published
|
|
1PC8
 
 | | Crystal Structure of a novel form of mistletoe lectin from Himalayan Viscum album L. at 3.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Himalayan mistletoe ribosome-inactivating protein, ... | | Authors: | Mishra, V, Ethayathulla, A.S, Paramasivam, M, Singh, G, Yadav, S, Kaur, P, Sharma, R.S, Babu, C.R, Singh, T.P. | | Deposit date: | 2003-05-16 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a novel ribosome-inactivating protein from a hemi-parasitic plant inhabiting the northwestern Himalayas.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5ZVJ
 
 | | Crystal structure of HtrA1 from Mycobacterium tuberculosis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Serine protease | | Authors: | Khundrakpam, H.S, Yadav, S, Kumar, D, Biswal, B.K. | | Deposit date: | 2018-05-10 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of an essential high-temperature requirement protein HtrA1 (Rv1223) from Mycobacterium tuberculosis reveals its unique features.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7AKT
 
 | | CrPetF variant - A39G_A41V | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kurisu, G, Ohnishi, Y, Engelbrecht, V, Happe, T. | | Deposit date: | 2020-10-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Ferredoxin 2.0: an electron transfer protein designed into a photosystem I-driven hydrogenase
To Be Published
|
|
7QHF
 
 | | [FeFe]-hydrogenase I from Clostridium pasteurianum (CpI), variant G302S | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Brocks, C, Duan, J, Hofmann, E, Happe, T. | | Deposit date: | 2021-12-12 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A Dynamic Water Channel Affects O 2 Stability in [FeFe]-Hydrogenases.
Chemsuschem, 2023
|
|
8QM3
 
 | |
8PVM
 
 | | formaldehyde-inhibited [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant C299D | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FORMYL GROUP, ... | | Authors: | Duan, J, Hofmann, E, Happe, T. | | Deposit date: | 2023-07-18 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Insights into the Molecular Mechanism of Formaldehyde Inhibition of [FeFe]-Hydrogenases.
J.Am.Chem.Soc., 145, 2023
|
|
6M0T
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin derivative (CL-2) | | Descriptor: | (3R)-3-[(R)-[(2R,6S)-6-methyloxan-2-yl]-oxidanyl-methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase | | Authors: | Babbar, P, Sharma, A, Manickam, Y. | | Deposit date: | 2020-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Cladosporin-Based Inhibitors of Malaria Parasites.
Acs Infect Dis., 7, 2021
|
|
8CJY
 
 | | [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant S357T | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Brocks, C, Duan, J, Hofmann, E, Happe, T. | | Deposit date: | 2023-02-13 | | Release date: | 2023-10-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Dynamic Water Channel Affects O 2 Stability in [FeFe]-Hydrogenases.
Chemsuschem, 17, 2024
|
|
1U4J
 
 | | Crystal structure of a carbohydrate induced dimer of group I phospholipase A2 from Bungarus caeruleus at 2.1 A resolution | | Descriptor: | ACETIC ACID, CHLORIDE ION, SODIUM ION, ... | | Authors: | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Betzel, C, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-07-26 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of a carbohydrate induced homodimer of phospholipase A(2) from Bungarus caeruleus at 2.1A resolution
J.Struct.Biol., 149, 2005
|
|
6LVV
 
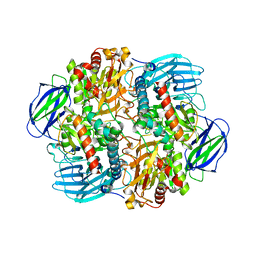 | | N, N-dimethylformamidase | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N,N-dimethylformamidase large subunit, ... | | Authors: | Arya, C.K, Ramaswamy, S, Kutti, R.V, Gurunath, R. | | Deposit date: | 2020-02-05 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5XEP
 
 | | Crystal structure of BRP39, a chitinase-like protein, at 2.6 Angstorm resolution | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-3-like protein 1 | | Authors: | Mohanty, A.K, Fisher, A.J, Choudhary, S, Kaushik, J.K. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of BRP39, a signalling glycoprotein expressed during mammary gland apoptosis.
To be published
|
|
