4G0L
 
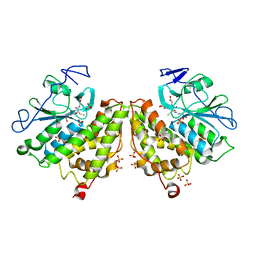 | | Glutathionyl-hydroquinone Reductase, YqjG, of E.coli complexed with GSH | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
6PXS
 
 | | Crystal structure of iminodiacetate oxidase (IdaA) from Chelativorans sp. BNC1 | | Descriptor: | FAD dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Jun, S.Y, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2019-07-26 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.836 Å) | | Cite: | Structural and biochemical characterization of iminodiacetate oxidase from Chelativorans sp. BNC1.
Mol.Microbiol., 112, 2019
|
|
5DQP
 
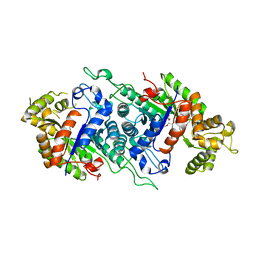 | | EDTA monooxygenase (EmoA) from Chelativorans sp. BNC1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, EDTA monooxygenase, SULFATE ION | | Authors: | Jun, S.Y, Youn, B, Xun, L, Kang, C, Lewis, K.M. | | Deposit date: | 2015-09-15 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Structural and biochemical characterization of EDTA monooxygenase and its physical interaction with a partner flavin reductase.
Mol.Microbiol., 100, 2016
|
|
6WM6
 
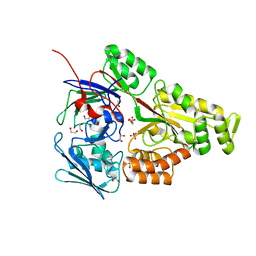 | | Periplasmic EDTA-binding protein EppA, tetragonal | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein, family 5, ... | | Authors: | Lewis, K.M, Sattler, S.A, Greene, C.L, Xun, L, Kang, C. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Structural Basis of the Binding of Various Aminopolycarboxylates by the Periplasmic EDTA-Binding Protein EppA from Chelativorans sp. BNC1.
Int J Mol Sci, 21, 2020
|
|
6WM7
 
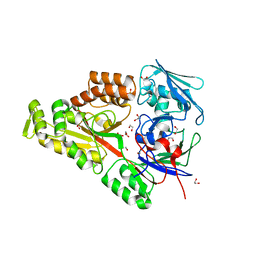 | | Periplasmic EDTA-binding protein EppA, orthorhombic | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein, family 5, ... | | Authors: | Lewis, K.M, Greene, C.L, Sattler, S.A, Xun, L, Kang, C. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The Structural Basis of the Binding of Various Aminopolycarboxylates by the Periplasmic EDTA-Binding Protein EppA from Chelativorans sp. BNC1.
Int J Mol Sci, 21, 2020
|
|
4HUZ
 
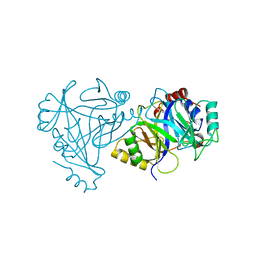 | | 2,6-Dichloro-p-hydroquinone 1,2-Dioxygenase | | Descriptor: | 2,6-dichloro-p-hydroquinone 1,2-dioxygenase, FE (III) ION, SULFATE ION | | Authors: | Hayes, R.P, Nissen, M.S, Green, A.R, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2012-11-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of 2,6-dichloro-p-hydroquinone 1,2-dioxygenase (PcpA) from Sphingobium chlorophenolicum, a new type of aromatic ring-cleavage enzyme.
Mol.Microbiol., 88, 2013
|
|
4LBP
 
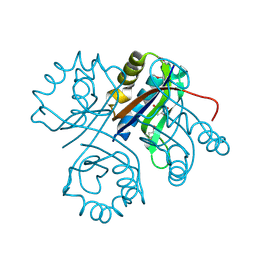 | | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) from Burkholderia phenoliruptrix AC1100: Complex with 2,5-dihydroxybenzoquinone | | Descriptor: | 2,5-dihydroxycyclohexa-2,5-diene-1,4-dione, 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) | | Authors: | Hayes, R.P, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Catalytic Mechanism of 5-Chlorohydroxyhydroquinone Dehydrochlorinase from the YCII Superfamily of Largely Unknown Function.
J.Biol.Chem., 288, 2013
|
|
4YSL
 
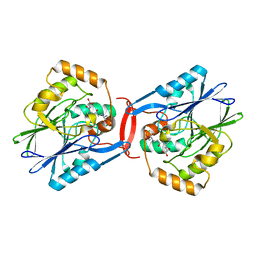 | | Crystal structure of SdoA from Pseudomonas putida in complex with glutathione | | Descriptor: | Beta-lactamase domain protein, FE (III) ION, GLUTATHIONE | | Authors: | Sattler, S.A, Wang, X, DeHan, P.J, Xun, L, Kang, C. | | Deposit date: | 2015-03-17 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4618 Å) | | Cite: | Characterizations of Two Bacterial Persulfide Dioxygenases of the Metallo-beta-lactamase Superfamily.
J.Biol.Chem., 290, 2015
|
|
4YSB
 
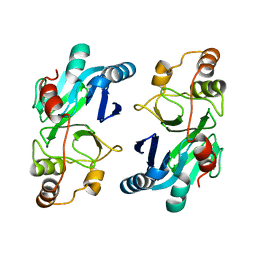 | | Crystal structure of ETHE1 from Myxococcus xanthus | | Descriptor: | FE (III) ION, Metallo-beta-lactamase family protein | | Authors: | Sattler, S.A, Wang, X, DeHan, P.J, Xun, L, Kang, C. | | Deposit date: | 2015-03-17 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5015 Å) | | Cite: | Characterizations of Two Bacterial Persulfide Dioxygenases of the Metallo-beta-lactamase Superfamily.
J.Biol.Chem., 290, 2015
|
|
4YSK
 
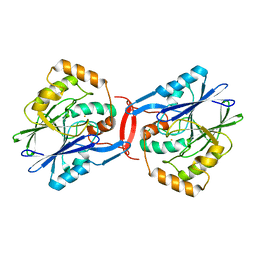 | | Crystal structure of apo-form SdoA from Pseudomonas putida | | Descriptor: | Beta-lactamase domain protein, FE (III) ION | | Authors: | Sattler, S.A, Wang, X, DeHan, P.J, Xun, L, Kang, C. | | Deposit date: | 2015-03-17 | | Release date: | 2015-06-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.466 Å) | | Cite: | Characterizations of Two Bacterial Persulfide Dioxygenases of the Metallo-beta-lactamase Superfamily.
J.Biol.Chem., 290, 2015
|
|
4FQU
 
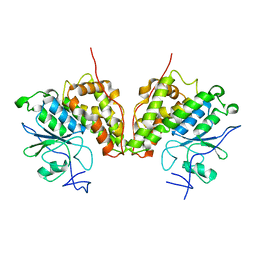 | |
4G0K
 
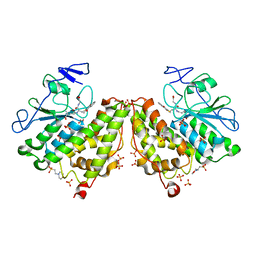 | | Glutathionyl-hydroquinone reductase, YqjG, of E.coli complexed with GS-menadione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-gamma-glutamyl-S-(3-methyl-1,4-dioxo-1,4-dihydronaphthalen-2-yl)-L-cysteinylglycine, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.557 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4G5E
 
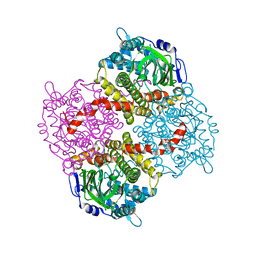 | | 2,4,6-Trichlorophenol 4-monooxygenase | | Descriptor: | 2,4,6-Trichlorophenol 4-monooxygenase | | Authors: | Hayes, R.P, Webb, B.N, Subramanian, A.K, Nissen, M, Popchock, A, Xun, L, Kang, C. | | Deposit date: | 2012-07-17 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Catalytic Differences between Two FADH(2)-Dependent Monooxygenases: 2,4,5-TCP 4-Monooxygenase (TftD) from Burkholderia cepacia AC1100 and 2,4,6-TCP 4-Monooxygenase (TcpA) from Cupriavidus necator JMP134.
Int J Mol Sci, 13, 2012
|
|
4G0I
 
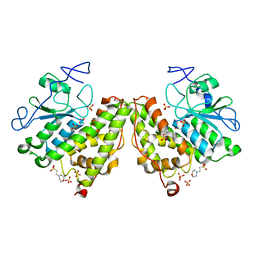 | | Glutathionyl-Hydroquinone Reductase, YqjG of Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, protein yqjG | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4LTM
 
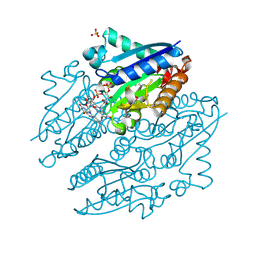 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
4LBH
 
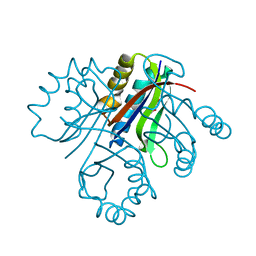 | | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) from Burkholderia phenoliruptrix AC1100: Apo-form | | Descriptor: | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG), DI(HYDROXYETHYL)ETHER | | Authors: | Hayes, R.P, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalytic Mechanism of 5-Chlorohydroxyhydroquinone Dehydrochlorinase from the YCII Superfamily of Largely Unknown Function.
J.Biol.Chem., 288, 2013
|
|
4LBI
 
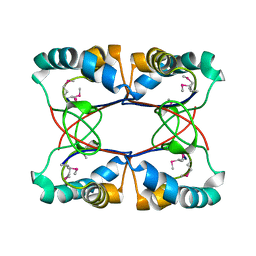 | |
4LTD
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - apo form | | Descriptor: | NADH-dependent FMN reductase, PHOSPHATE ION, SULFATE ION | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
4LTN
 
 | | Crystal structures of NADH:FMN oxidoreductase (EMOB) - FMN, NADH complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, NADH-dependent FMN reductase, ... | | Authors: | Nissen, M.S, Youn, B, Knowles, B.D, Ballinger, J.W, Jun, S, Belchik, S.M, Xun, L, Kang, C. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis.
J.Biol.Chem., 283, 2008
|
|
3S2I
 
 | | Crystal Structure of FurX NADH+:Furfuryl alcohol II | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
4RNS
 
 | | PcpR inducer binding domain (apo-form) | | Descriptor: | PCP degradation transcriptional activation protein | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-25 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
4RPO
 
 | | PcpR inducer binding domain (Complex with 2,4,6-trichlorophenol) | | Descriptor: | 2,4,6-trichlorophenol, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
3S1L
 
 | | Crystal Structure of Apo-form FurX | | Descriptor: | HEXAETHYLENE GLYCOL, ZINC ION, Zinc-containing alcohol dehydrogenase superfamily | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-15 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Furfural reduction mechanism of a zinc-dependent alcohol dehydrogenase from Cupriavidus necator JMP134.
Mol.Microbiol., 83, 2012
|
|
3S2E
 
 | | Crystal Structure of FurX NADH Complex 1 | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal Structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
4RPN
 
 | | PcpR inducer binding domain complex with pentachlorophenol | | Descriptor: | PCP degradation transcriptional activation protein, PENTACHLOROPHENOL | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
