4DBH
 
 | | Crystal structure of Cg1458 with inhibitor | | Descriptor: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION, OXALATE ION | | Authors: | Ran, T.T, Wang, W.W, Xu, D.Q, Gao, Y.Y. | | Deposit date: | 2012-01-15 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
4DBF
 
 | | Crystal structures of Cg1458 | | Descriptor: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION | | Authors: | Ran, T.T, Xu, D.Q, Wang, W.W, Gao, Y.Y, Wang, M.T. | | Deposit date: | 2012-01-15 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
1OQE
 
 | | Crystal structure of sTALL-1 with BAFF-R | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
1OQD
 
 | | Crystal structure of sTALL-1 and BCMA | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
1IAS
 
 | | CYTOPLASMIC DOMAIN OF UNPHOSPHORYLATED TYPE I TGF-BETA RECEPTOR CRYSTALLIZED WITHOUT FKBP12 | | Descriptor: | SULFATE ION, TGF-BETA RECEPTOR TYPE I | | Authors: | Huse, M, Muir, T.W, Chen, Y.-G, Kuriyan, J, Massague, J. | | Deposit date: | 2001-03-23 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The TGF beta receptor activation process: an inhibitor- to substrate-binding switch.
Mol.Cell, 8, 2001
|
|
8J9O
 
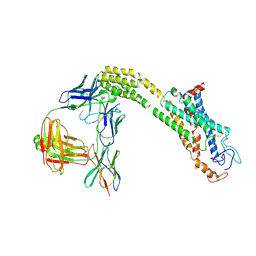 | | Cryo-EM structure of inactive FZD1 | | Descriptor: | Frizzled-1,Soluble cytochrome b562, anti-BRIL Fab Heavy Chain, anti-BRIL Fab Light Chain, ... | | Authors: | Lin, X, Xu, F. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8J9N
 
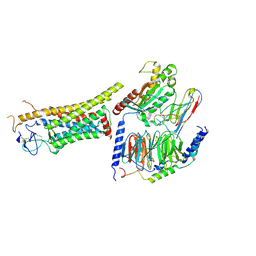 | | Gq bound FZD1 in ligand-free state | | Descriptor: | Frizzled-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lin, X, Xu, F. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8JH7
 
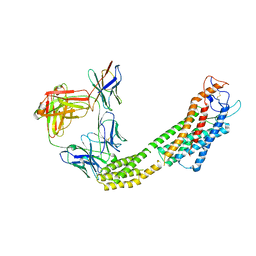 | | FZD6 in inactive state | | Descriptor: | Frizzled-6,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-22 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8JHI
 
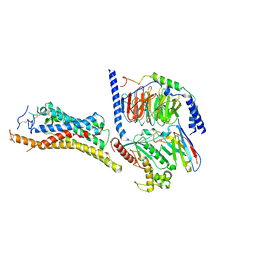 | | FZD3-Gs complex | | Descriptor: | Frizzled-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8JHC
 
 | | FZD3 in inactive state | | Descriptor: | Frizzled-3,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8JHB
 
 | | FZD6 Gs complex | | Descriptor: | Frizzled-6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8GXP
 
 | | Complex structure of RORgama with betulinic acid | | Descriptor: | Betulinic acid, Nuclear receptor ROR-gamma | | Authors: | Zhang, X.L, Xu, C, Bai, F. | | Deposit date: | 2022-09-20 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery, structural optimization, and anti-tumor bioactivity evaluations of betulinic acid derivatives as a new type of ROR gamma antagonists.
Eur.J.Med.Chem., 257, 2023
|
|
6LHC
 
 | | The cryo-EM structure of coxsackievirus A16 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHL
 
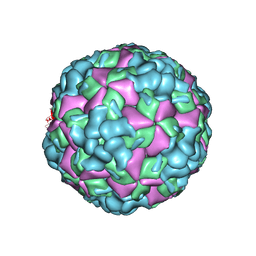 | | The cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHB
 
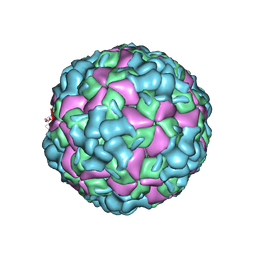 | | The cryo-EM structure of coxsackievirus A16 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHP
 
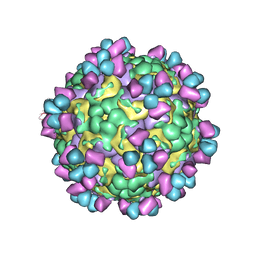 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 14B10 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHO
 
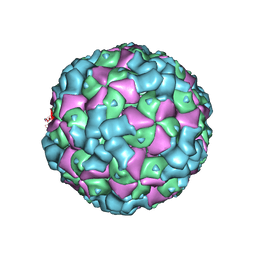 | | The cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHK
 
 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHA
 
 | | The cryo-EM structure of coxsackievirus A16 mature virion | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHQ
 
 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab NA9D7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHT
 
 | | Localized reconstruction of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, heavy chain variable region of Fab 18A7, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
5DRB
 
 | | Crystal structure of WNK1 in complex with WNK463 | | Descriptor: | N-tert-butyl-1-(1-{5-[5-(trifluoromethyl)-1,3,4-oxadiazol-2-yl]pyridin-2-yl}piperidin-4-yl)-1H-imidazole-5-carboxamide, Serine/threonine-protein kinase WNK1 | | Authors: | Kohls, D, Xie, X. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Small-molecule WNK inhibition regulates cardiovascular and renal function.
Nat.Chem.Biol., 12, 2016
|
|
8DEY
 
 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
8STI
 
 | | human STING with agonist XMT-1616 | | Descriptor: | 3-[(2E)-4-{5-carbamoyl-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-(3-hydroxypropoxy)-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-3H-imidazo[4,5-b]pyridine-6-carboxamide, Stimulator of interferon genes protein | | Authors: | Duvall, J.R, Bukhalid, R.A. | | Deposit date: | 2023-05-10 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and Optimization of a STING Agonist Platform for Application in Antibody Drug Conjugates.
J.Med.Chem., 66, 2023
|
|
8STH
 
 | | human STING with diABZI agonist 15 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-(3-hydroxypropoxy)-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Duvall, J.R, Bukhalid, R.A. | | Deposit date: | 2023-05-10 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Optimization of a STING Agonist Platform for Application in Antibody Drug Conjugates.
J.Med.Chem., 66, 2023
|
|
