6VC5
 
 | |
6UAG
 
 | |
6UG4
 
 | |
6UHS
 
 | |
6UHH
 
 | | Crystal Structure of Human RYR Receptor 3 ( 848-1055) in Complex with ATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, R, Kim, Y, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-09-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.138 Å) | | Cite: | Crystal Structure of Human RYR Receptor 3 ( 848-1055) in Complex with ATP
To Be Published
|
|
6UHA
 
 | |
6UHB
 
 | |
6UAM
 
 | |
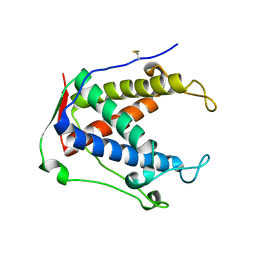
6UHE
 
 | |
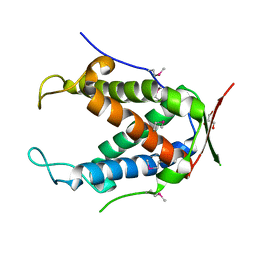
6UHI
 
 | |
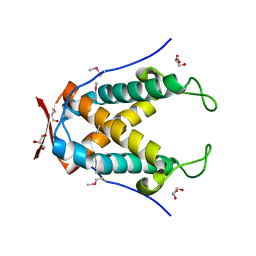
6UG5
 
 | |
1RLJ
 
 | | Structural Genomics, a Flavoprotein NrdI from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, IODIDE ION, NrdI protein | | Authors: | Wu, R, Zhang, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1.5A crystal structure of a thioredoxin-like protein NrdI from Bacillus subtilis
To be Published
|
|
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
8SPR
 
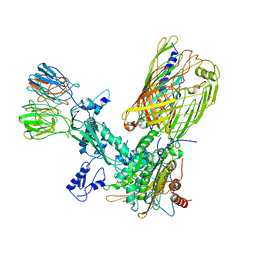 | | The cryo-EM structure of the EcBAM/EspP(beta1-12) complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R, Noinaj, N. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | BAM orchestrates OMP biogenesis using a beta-templating mechanism
To Be Published
|
|
8SQB
 
 | | The cryo-EM structure of the EcBAM/EspP(beta7-12) complex | | Descriptor: | Maltodextrin-binding protein,EspP(b7-12), Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R, Noinaj, N. | | Deposit date: | 2023-05-04 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | BAM orchestrates OMP biogenesis using a beta-templating mechanism
To Be Published
|
|
8SQA
 
 | | The cryo-EM structure of the EcBAM/EspP(beta8-12) complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Autotransporter protein EspP translocator, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R, Noinaj, N. | | Deposit date: | 2023-05-04 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | BAM orchestrates OMP biogenesis using a beta-templating mechanism
To Be Published
|
|
2PPW
 
 | |
1XFK
 
 | |
6VC6
 
 | | 2.1 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Mn2+ | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-phospho-alpha-glucosidase, GLYCEROL, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2019-12-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Mn2+
To Be Published
|
|
3K32
 
 | |
3BK5
 
 | |
3C85
 
 | |
3U27
 
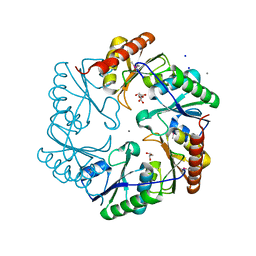 | | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b | | Descriptor: | CALCIUM ION, GLYCEROL, Microcompartments protein, ... | | Authors: | Wu, R, Gu, M, Kerfeld, C.A, Salmeen, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-01 | | Release date: | 2012-02-08 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b
To be Published
|
|
2OT9
 
 | |
2P7J
 
 | |
