8B16
 
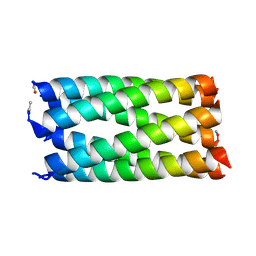 | |
8B15
 
 | |
8B45
 
 | | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4 | | Descriptor: | 1,2-ETHANEDIOL, CC-Tri-(UbUc)4, SODIUM ION, ... | | Authors: | Kumar, P, Martin, F.J.O, Dawson, W.M, Zieleniewski, F, Woolfson, D.N. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4
To Be Published
|
|
7QWC
 
 | |
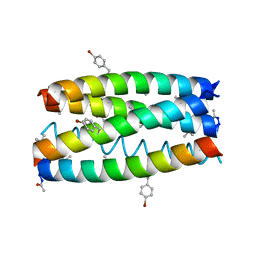
7QWE
 
 | |
7QWB
 
 | |
7QWA
 
 | |
7QWD
 
 | |
1EYM
 
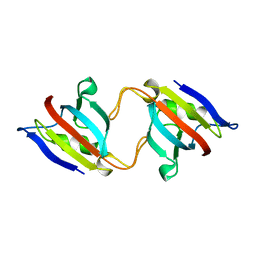 | | FK506 BINDING PROTEIN MUTANT, HOMODIMERIC COMPLEX | | Descriptor: | FK506 BINDING PROTEIN | | Authors: | Rollins, C.T, Rivera, V.M, Woolfson, D.N, Keenan, T, Hatada, M, Adams, S.E, Andrade, L.J, Yaeger, D, van Schravendijk, M.R, Holt, D.A, Gilman, M, Clackson, T. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ligand-reversible dimerization system for controlling protein-protein interactions.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5LO4
 
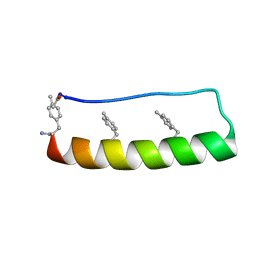 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPa-CH3 | | Authors: | Baker, E.G, Williams, C, Hudson, K.L, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5LO2
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaTyr | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5LO3
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaOMe | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
2G0K
 
 | |
2G0L
 
 | | Solution Structure of Neocarzinostatin Apo-Protein with bound Flavone | | Descriptor: | 2-PHENYL-4H-CHROMEN-4-ONE, NEOCARZINOSTATIN | | Authors: | Muskett, F.W, Stoneman, R.G, Caddick, S, Woolfson, D.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Synthetic Ligands for Apo-Neocarzinostatin
J.Am.Chem.Soc., 128, 2006
|
|
1SIF
 
 | | Crystal structure of a multiple hydrophobic core mutant of ubiquitin | | Descriptor: | ubiquitin | | Authors: | Benitez-Cardoza, C.G, Stott, K, Hirshberg, M, Went, H.M, Woolfson, D.N, Jackson, S.E. | | Deposit date: | 2004-02-29 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exploring sequence/folding space: folding studies on multiple hydrophobic core mutants of ubiquitin
Biochemistry, 43, 2004
|
|
4KVU
 
 | | Crystal structure of a 6-helix coiled coil CC-Hex-L17C-W224BF | | Descriptor: | 6-helix coiled coil CC-Hex-L17C-W224BF, GLYCEROL | | Authors: | Burton, A.J, Agnew, C, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-21 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Accessibility, Reactivity, and Selectivity of Side Chains within a Channel of de Novo Peptide Assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4KVV
 
 | | Crystal structure of an alkylated Cys mutant of CC-Hex | | Descriptor: | 6-HELIX COILED COIL CC-HEX-L24C PEPTIDE with an alkylated Cys mutation | | Authors: | Burton, A.J, Agnew, C, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-21 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Accessibility, Reactivity, and Selectivity of Side Chains within a Channel of de Novo Peptide Assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4KVT
 
 | | Crystal structure of a 6-helix coiled coil CC-Hex-L24C | | Descriptor: | 6-helix coiled coil CC-Hex-L24C peptide | | Authors: | Burton, A.J, Agnew, C, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-21 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accessibility, Reactivity, and Selectivity of Side Chains within a Channel of de Novo Peptide Assembly.
J.Am.Chem.Soc., 135, 2013
|
|
8OYK
 
 | | Coiled-Coil Domain of Human STIL, L736E Mutant | | Descriptor: | CHLORIDE ION, Isoform 2 of SCL-interrupting locus protein | | Authors: | Martin, F.J.O, Shamir, M, Woolfson, D.N, Friedler, A. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Mechanism of STIL Coiled-Coil Domain Oligomerization.
Int J Mol Sci, 24, 2023
|
|
8OYL
 
 | | Coiled-Coil Domain of Human STIL, Q729L Mutant | | Descriptor: | CADMIUM ION, CHLORIDE ION, SCL-interrupting locus protein, ... | | Authors: | Martin, F.J.O, Shamir, M, Woolfson, D.N, Friedler, A. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular Mechanism of STIL Coiled-Coil Domain Oligomerization.
Int J Mol Sci, 24, 2023
|
|
6Q5I
 
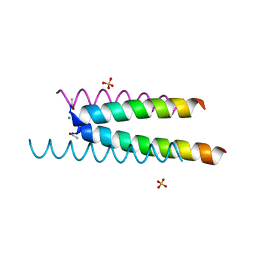 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24E | | Descriptor: | AMMONIUM ION, CC-Hex*-L24E, SULFATE ION | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5L
 
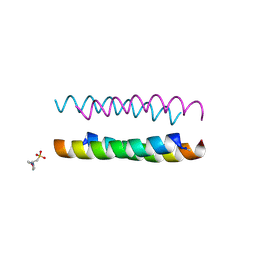 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24H | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CC-Hex*-L24H | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5P
 
 | | Crystal structure of a CC-Hex mutant that forms a parallel six-helix coiled coil CC-Hex*-II | | Descriptor: | CC-Hex*-II, GLYCEROL, PHOSPHATE ION | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5O
 
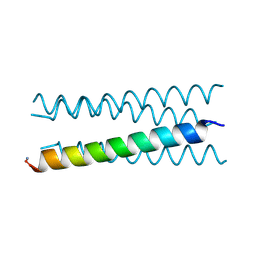 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-LL | | Descriptor: | CC-Hex*-LL | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
8BFD
 
 | | Racemic structure of PK-7 (310HD-U2U5) | | Descriptor: | 310HD-U2U5, D-310HD-U2U5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, P, Paterson, N.G, Woolfson, D.N. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of discrete, stable 3 10 -helix peptide assemblies.
Nature, 607, 2022
|
|
