2J4M
 
 | | Double dockerin from Piromyces equi Cel45A | | Descriptor: | ENDOGLUCANASE 45A | | Authors: | Nagy, T, Tunnicliffe, R.B, Higgins, L.D, Walters, C, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2006-09-01 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Double Dockerin from the Cellulosome of the Anaerobic Fungus Piromyces Equi.
J.Mol.Biol., 373, 2007
|
|
2J52
 
 | | Solution Structure of GB1 domain Protein G and low and high pressure. | | Descriptor: | IMMUNOGLOBULIN G-BINDING PROTEIN G | | Authors: | Wilton, D.J, Tunnicliffe, R.B, Kamatari, Y.O, Akasaka, K, Williamson, M.P. | | Deposit date: | 2006-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pressure-Induced Changes in the Solution Structure of the Gb1 Domain of Protein G.
Proteins, 71, 2008
|
|
2J4N
 
 | | Double dockerin from Piromyces equi Cel45A | | Descriptor: | ENDOGLUCANASE 45A | | Authors: | Nagy, T, Tunnicliffe, R.B, Higgins, L.D, Walters, C, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2006-09-01 | | Release date: | 2007-09-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Double Dockerin from the Cellulosome of the Anaerobic Fungus Piromyces Equi.
J.Mol.Biol., 373, 2007
|
|
2J53
 
 | | Solution Structure of GB1 domain Protein G and low and high pressure. | | Descriptor: | IMMUNOGLOBULIN G-BINDING PROTEIN G | | Authors: | Wilton, D.J, Tunnicliffe, R.B, Kamatari, Y.O, Akasaka, K, Williamson, M.P. | | Deposit date: | 2006-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pressure-Induced Changes in the Solution Structure of the Gb1 Domain of Protein G.
Proteins, 71, 2008
|
|
2MKX
 
 | |
2NRG
 
 | |
6RK4
 
 | | Lysostaphin SH3b P4-G5 complex, synchrotron dataset | | Descriptor: | (2~{R})-2-[[(2~{S})-2-[[(4~{R})-5-azanyl-4-[[(2~{S})-2-azanylpropanoyl]amino]-5-oxidanylidene-pentanoyl]amino]-6-[2-[2-[2-[2-(2-azanylethanoylamino)ethanoylamino]ethanoylamino]ethanoylamino]ethanoylamino]hexanoyl]amino]propanoic acid, 1,2-ETHANEDIOL, Lysostaphin | | Authors: | Walters-Morgan, H, Lovering, A.L. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Two-site recognition of Staphylococcus aureus peptidoglycan by lysostaphin SH3b.
Nat.Chem.Biol., 16, 2020
|
|
6RJE
 
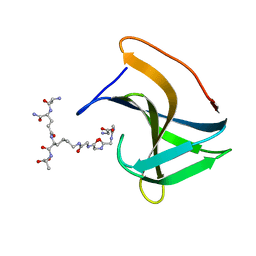 | | Lysostaphin SH3b P4-G5 complex, homesource dataset | | Descriptor: | (2~{R})-2-[[(2~{S})-2-[[(4~{R})-5-azanyl-4-[[(2~{S})-2-azanylpropanoyl]amino]-5-oxidanylidene-pentanoyl]amino]-6-[2-[2-[2-[2-(2-azanylethanoylamino)ethanoylamino]ethanoylamino]ethanoylamino]ethanoylamino]hexanoyl]amino]propanoic acid, Lysostaphin | | Authors: | Walters-Morgan, H, Lovering, A.L. | | Deposit date: | 2019-04-26 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two-site recognition of Staphylococcus aureus peptidoglycan by lysostaphin SH3b.
Nat.Chem.Biol., 16, 2020
|
|
1G1N
 
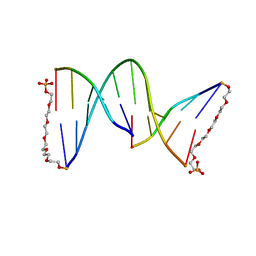 | | NICKED DECAMER DNA WITH PEG6 TETHER, NMR, 30 STRUCTURES | | Descriptor: | 5'-D(*GP*TP*CP*GP*C)-3', 5'-D(P*GP*CP*GP*AP*CP*AP*AP*CP*GP*C)-3', 5'-D(P*GP*CP*GP*TP*T)-3', ... | | Authors: | Bocian, W, Kozerski, L, Mazurek, A.P, Kawecki, R. | | Deposit date: | 2000-10-13 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A nicked duplex decamer DNA with a PEG(6) tether.
Nucleic Acids Res., 29, 2001
|
|
2RSU
 
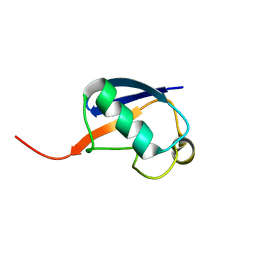 | | Alternative structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Kitazawa, S, Kameda, T, Yagi-Utsumi, M, Kato, K, Kitahara, R. | | Deposit date: | 2012-06-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Q41N Variant of Ubiquitin as a Model for the Alternatively Folded N2 State of Ubiquitin
Biochemistry, 52, 2013
|
|
2WZ8
 
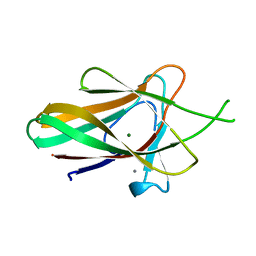 | | Family 35 carbohydrate binding module from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSOME PROTEIN DOCKERIN TYPE I, MAGNESIUM ION | | Authors: | Gloster, T.M, Davies, G.J, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2009-11-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Signature Active Site Architectures Illuminate the Molecular Basis for Ligand Specificity in Family 35 Carbohydrate Binding Module .
Biochemistry, 49, 2010
|
|
2KNV
 
 | |
