4CC5
 
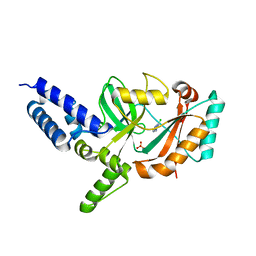 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
1FOC
 
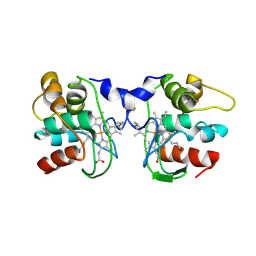 | | Cytochrome C557: improperly folded thermus thermophilus C552 | | Descriptor: | CYTOCHROME RC557, HEME C | | Authors: | McRee, D.E, Williams, P.A, Fee, J.A, Bren, K.L. | | Deposit date: | 2000-08-27 | | Release date: | 2000-11-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recombinant cytochrome rC557 obtained from Escherichia coli cells expressing a truncated Thermus thermophilus cycA gene. Heme inversion in an improperly matured protein
J.Biol.Chem., 276, 2001
|
|
4CXJ
 
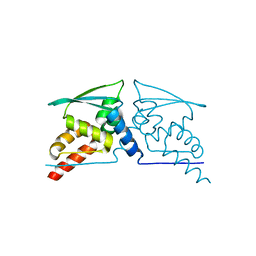 | | BTB domain of KEAP1 C151W mutant | | Descriptor: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Cleasby, A, Yon, J, Day, P.J, Richardson, C, Tickle, I.J, Williams, P.A, Callahan, J.F, Carr, R, Concha, N, Kerns, J.K, Qi, H, Sweitzer, T, Ward, P, Davies, T.G. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Btb Domain of Keap1 and its Interaction with the Triterpenoid Antagonist Cddo.
Plos One, 9, 2014
|
|
4CXI
 
 | | BTB domain of KEAP1 | | Descriptor: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Cleasby, A, Yon, J, Day, P.J, Richardson, C, Tickle, I.J, Williams, P.A, Callahan, J.F, Carr, R, Concha, N, Kerns, J.K, Qi, H, Sweitzer, T, Ward, P, Davies, T.G. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Btb Domain of Keap1 and its Interaction with the Triterpenoid Antagonist Cddo.
Plos One, 9, 2014
|
|
4CXT
 
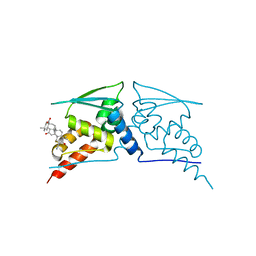 | | BTB domain of KEAP1 in complex with CDDO | | Descriptor: | (13alpha,18alpha)-2-cyano-3-hydroxy-12-oxooleana-2,9(11)-dien-28-oic acid, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Cleasby, A, Yon, J, Day, P.J, Richardson, C, Tickle, I.J, Williams, P.A, Callahan, J.F, Carr, R, Concha, N, Kerns, J.K, Qi, H, Sweitzer, T, Ward, P, Davies, T.G. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of the Btb Domain of Keap1 and its Interaction with the Triterpenoid Antagonist Cddo.
Plos One, 9, 2014
|
|
5FPS
 
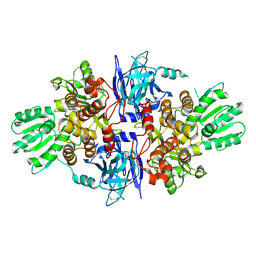 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 3-aminobenzene-1,2-dicarboxylic acid (AT1246) in an alternate binding site. | | Descriptor: | 3-AMINOBENZENE-1,2-DICARBOXYLIC ACID, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPR
 
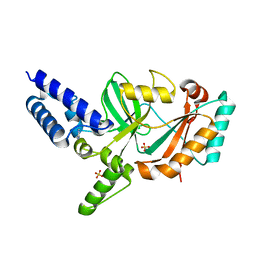 | | Structure of Bacterial DNA Ligase with small-molecule ligand pyrimidin-2-amine (AT371) in an alternate binding site. | | Descriptor: | DNA LIGASE, PYRIMIDIN-2-AMINE, SULFATE ION | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPO
 
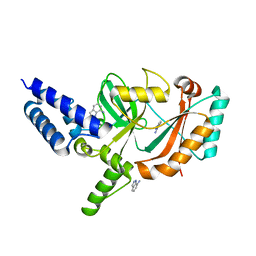 | | Structure of Bacterial DNA Ligase with small-molecule ligand 1H- indazol-7-amine (AT4213) in an alternate binding site. | | Descriptor: | 1H-indazol-7-amine, DNA LIGASE | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPT
 
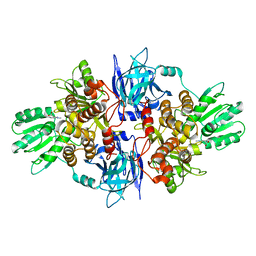 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 2-(1-methyl-1H-indol-3-yl)acetic acid (AT3437) in an alternate binding site. | | Descriptor: | (1-methyl-1H-indol-3-yl)acetic acid, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Pathuri, P, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2CAG
 
 | | CATALASE COMPOUND II | | Descriptor: | CATALASE COMPOUND II, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gouet, P, Jouve, H.M, Hajdu, J. | | Deposit date: | 1996-06-12 | | Release date: | 1996-12-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ferryl intermediates of catalase captured by time-resolved Weissenberg crystallography and UV-VIS spectroscopy.
Nat.Struct.Biol., 3, 1996
|
|
1OD1
 
 | | Endothiapepsin PD135,040 complex | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-12 | | Last modified: | 2012-11-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The Structure of Endothiapepsin Complexed with the Gem-Diol Inhibitor Pd-135,040 at 1.37 A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5FPY
 
 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 5-bromo-1-methyl-1H-indole-2-carboxylic acid (AT21457) in an alternate binding site. | | Descriptor: | 5-bromo-1-methyl-1H-indole-2-carboxylic acid, SERINE PROTEASE NS3 | | Authors: | Davies, T.G, Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1H9X
 
 | | Cytochrome cd1 Nitrite Reductase, reduced form | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CYTOCHROME CD1 NITRITE REDUCTASE, HEME C, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-03-23 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of an Alternative Form of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase
J.Biol.Chem., 276, 2001
|
|
1HCM
 
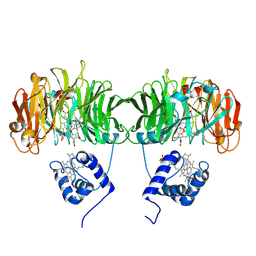 | |
1H6N
 
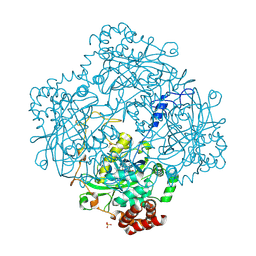 | | Formation of a tyrosyl radical intermediate in Proteus mirabilis catalase by directed mutagenesis and consequences for nucleotide reactivity | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Sainz, G, Jaquinod, M, Gagnon, J, Jouve, H.M. | | Deposit date: | 2001-06-20 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High Resolution Structure and Biochemical Properties of a Recombinant Proteus Mirabilis Catalase Depleted in Iron.
Proteins: Struct.,Funct., Genet., 50, 2003
|
|
1H9Y
 
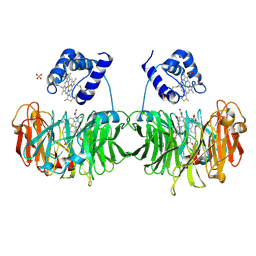 | |
1HJ3
 
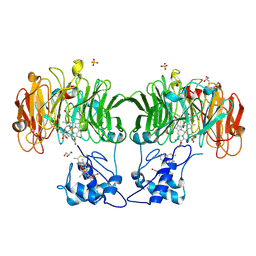 | | Cytochrome cd1 Nitrite Reductase, dioxygen complex | | Descriptor: | GLYCEROL, HEME C, HEME D, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the bound dioxygen species in the cytochrome oxidase reaction of cytochrome cd1 nitrite reductase.
J. Biol. Chem., 276, 2001
|
|
1HJ5
 
 | | Cytochrome cd1 Nitrite Reductase, reoxidised enzyme | | Descriptor: | GLYCEROL, HEME C, HEME D, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of the bound dioxygen species in the cytochrome oxidase reaction of cytochrome cd1 nitrite reductase.
J. Biol. Chem., 276, 2001
|
|
1HJ4
 
 | |
1N6B
 
 | | Microsomal Cytochrome P450 2C5/3LVdH Complex with a dimethyl derivative of sulfaphenazole | | Descriptor: | 4-METHYL-N-METHYL-N-(2-PHENYL-2H-PYRAZOL-3-YL)BENZENESULFONAMIDE, Cytochrome P450 2C5, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wester, M.R, Johnson, E.F, Marques-Soares, C, Dansette, P.M, Mansuy, D, Stout, C.D. | | Deposit date: | 2002-11-09 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Substrate Complex of Mammalian Cytochrome P450 2C5 at 2.3 A Resolution: Evidence
for Multiple Substrate Binding Modes
Biochemistry, 42, 2003
|
|
1DY7
 
 | | Cytochrome cd1 Nitrite Reductase, CO complex | | Descriptor: | CARBON MONOXIDE, GLYCEROL, HEME C, ... | | Authors: | Sjogren, T, Svensson-Ek, M, Hajdu, J, Brzezinski, P. | | Deposit date: | 2000-01-28 | | Release date: | 2000-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proton-Coupled Structural Changes Upon Binding of Carbon Monoxide to Cytochrome Cd(1): A Combined Flash Photolysis and X-Ray Crystallography Study
Biochemistry, 39, 2000
|
|
1E2R
 
 | | CYTOCHROME CD1 NITRITE REDUCTASE, REDUCED AND CYANIDE BOUND | | Descriptor: | CYANIDE ION, GLYCEROL, HEME C, ... | | Authors: | Fulop, V. | | Deposit date: | 2000-05-24 | | Release date: | 2000-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-Ray Crystallographic Study of Cyanide Binding Provides Insights Into the Structure-Function Relationship for Cytochrome Cd1 Nitrite Reductase from Paracoccus Pantotrophus.
J.Biol.Chem., 275, 2000
|
|
1E93
 
 | | High resolution structure and biochemical properties of a recombinant catalase depleted in iron | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Sainz, G, Jaquinod, M, Gagnon, J, Jouve, H.M. | | Deposit date: | 2000-10-05 | | Release date: | 2000-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Resolution Structure and Biochemical Properties of a Recombinant Proteus Mirabilis Catalase Depleted in Iron.
Proteins: Struct.,Funct., Genet., 50, 2003
|
|
1H7K
 
 | | Formation of a tyrosyl radical intermediate in Proteus mirabilis catalase by directed mutagenesis and consequences for nucleotide reactivity | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Gambarelli, S, Gaillard, J, Sainz, G, Stojanoff, V, Jouve, H.M. | | Deposit date: | 2001-07-08 | | Release date: | 2004-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Formation of a Tyrosyl Radical Intermediate in Proteus Mirabilis Catalase by Directed Mutagenesis and Consequences for Nucleotide Reactivity.
Biochemistry, 40, 2001
|
|
