7E2B
 
 | | GDPD mutant complex from Pyrococcus furiosus DSM 3638 | | Descriptor: | CHOLINE ION, Glycerophosphodiester phosphodiesterase, MAGNESIUM ION, ... | | Authors: | Wang, Y.H, Wang, J, Wang, F.H. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Observation of the GPC production during hydrolysis of LPC by GDPD D60A from Pyrococcus furiosus DSM 3638
To Be Published
|
|
7E0V
 
 | | GDPD from Pyrococcus furiosus DSM 3638 | | Descriptor: | Glycerophosphodiester phosphodiesterase | | Authors: | Wang, Y.H, Wang, J, Wang, F.H. | | Deposit date: | 2021-01-28 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Bomgl, a monoacylglycerol lipase from marine Bacillus
To Be Published
|
|
7E2A
 
 | | GDPD mutant from Pyrococcus furiosus DSM 3638 | | Descriptor: | Glycerophosphodiester phosphodiesterase | | Authors: | Wang, Y.H, Wang, J, Wang, F.H. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of glycerophoshphodiester phospodiesterase D60A mutant from Pyrococcus furiosus DSM
To Be Published
|
|
7E04
 
 | |
7WU1
 
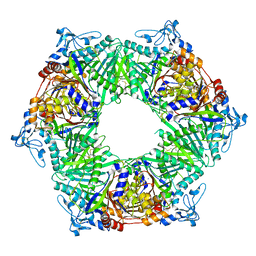 | | Crystal structure of phospholipase D from Moritella sp. JT01 | | Descriptor: | 1,2-ETHANEDIOL, Phospholipase D, SODIUM ION | | Authors: | Wang, Y.H, Mao, X.J, Wang, J, Wang, F.H. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of phospholipase D from Moritella sp. JT01
To Be Published
|
|
8HL4
 
 | |
8HL1
 
 | |
8HL2
 
 | |
8HL3
 
 | |
8HKV
 
 | |
8HL5
 
 | |
8HKY
 
 | |
8HKX
 
 | |
8HKZ
 
 | |
8HKU
 
 | |
6IDY
 
 | |
6L7N
 
 | |
7V3K
 
 | | crystal structure of MAJ1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y.H, Cui, R.G. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | crystal structure of MAJ1
To Be Published
|
|
5XK2
 
 | |
5XKS
 
 | |
7EOA
 
 | | HR-PETase from Bacterium HR29 | | Descriptor: | ETHANOL, Poly(Ethylene terephthalate) hydrolase | | Authors: | Wang, J, Wang, Y.H. | | Deposit date: | 2021-04-21 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of Bomgl, a monoacylglycerol lipase from marine Bacillus
To Be Published
|
|
8WQ4
 
 | |
8WQ2
 
 | |
8WKP
 
 | |
7SPR
 
 | |
