6WTW
 
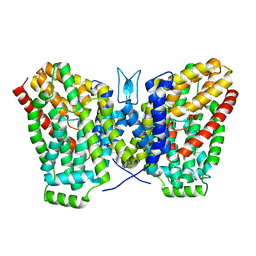 | | Structure of LaINDY crystallized in the presence of alpha-ketoglutarate and malate | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Cocco, N, Marden, J.J, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU4
 
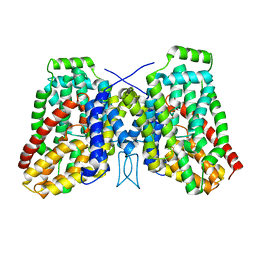 | | Structure of the LaINDY-alpha-ketoglutarate complex | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
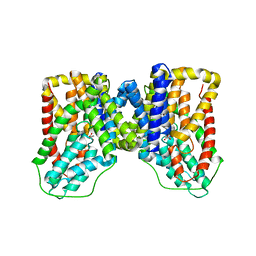
6WTX
 
 | |
6WU3
 
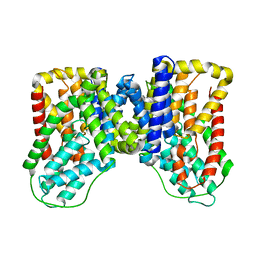 | |
4F35
 
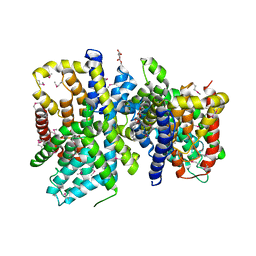 | | Crystal Structure of a bacterial dicarboxylate/sodium symporter | | Descriptor: | CITRIC ACID, SODIUM ION, Transporter, ... | | Authors: | Mancusso, R.L, Gregorio, G.G, Liu, Q, Wang, D.N. | | Deposit date: | 2012-05-08 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter.
Nature, 491, 2012
|
|
2QJU
 
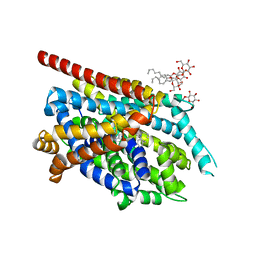 | | Crystal Structure of an NSS Homolog with Bound Antidepressant | | Descriptor: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, CHLORIDE ION, LEUCINE, ... | | Authors: | Zhou, Z, Karpowich, N.K, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2007-07-09 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake.
Science, 317, 2007
|
|
3TE0
 
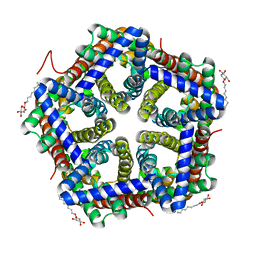 | | Crystal structure of HSC K148E | | Descriptor: | formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TDO
 
 | | Crystal structure of HSC at pH 9.0 | | Descriptor: | Putative formate/nitrite transporter, TETRAETHYLENE GLYCOL, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TDX
 
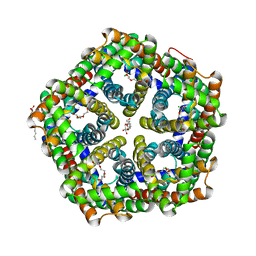 | | Crystal structure of HSC L82V | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, formate/nitrite transporter, ... | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TE2
 
 | | Crystal structure of HSC K16S | | Descriptor: | TETRAETHYLENE GLYCOL, formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TDS
 
 | | Crystal structure of HSC F194I | | Descriptor: | TETRAETHYLENE GLYCOL, formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TDR
 
 | | Crystal structure of HSC at pH 7.5 | | Descriptor: | TETRAETHYLENE GLYCOL, formate/nitrite transporter | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TE1
 
 | | Crystal structure of HSC T84A | | Descriptor: | TETRAETHYLENE GLYCOL, formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
3TDP
 
 | | Crystal structure of HSC at pH 4.5 | | Descriptor: | ZINC ION, formate/nitrite transporter, octyl beta-D-glucopyranoside | | Authors: | Czyzewski, B.K, Wang, D.-N. | | Deposit date: | 2011-08-11 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Identification and characterization of a bacterial hydrosulphide ion channel.
Nature, 483, 2012
|
|
2BHW
 
 | | PEA LIGHT-HARVESTING COMPLEX II AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6'S,9R,9'R,13R,13'S)-4',5'-DIDEHYDRO-5',6',7',8',9,9',10,10',11,11',12,12',13,13',14,14',15,15'-OCTADECAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Standfuss, J, Terwisscha van Scheltinga, A.C, Lamborghini, M, Kuehlbrandt, W. | | Deposit date: | 2005-01-19 | | Release date: | 2006-05-23 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms of Photoprotection and Nonphotochemical Quenching in Pea Light-Harvesting Complex at 2.5 A Resolution.
Embo J., 24, 2005
|
|
