2YK6
 
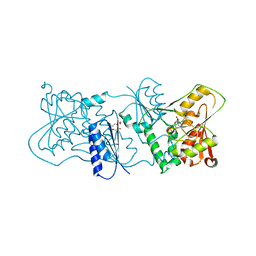 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CDP. | | Descriptor: | CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-DIPHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
1XNC
 
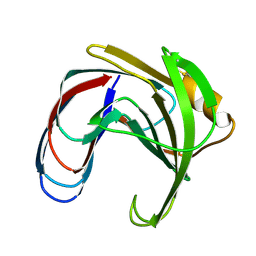 | |
3ECQ
 
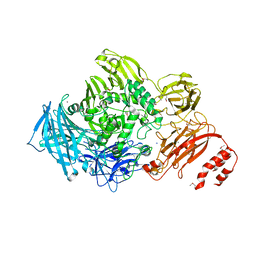 | | Endo-alpha-N-acetylgalactosaminidase from Streptococcus pneumoniae: SeMet structure | | Descriptor: | CALCIUM ION, Endo-alpha-N-acetylgalactosaminidase, GLYCEROL, ... | | Authors: | Caines, M.E.C, Zhu, H, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structural basis for T-antigen hydrolysis by Streptococcus pneumoniae: a target for structure-based vaccine design.
J.Biol.Chem., 283, 2008
|
|
2DRJ
 
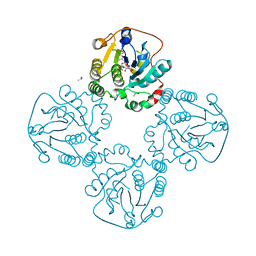 | |
2FNI
 
 | | PseC aminotransferase involved in pseudoaminic acid biosynthesis | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Cygler, M, Matte, A, Lunin, V.V, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
2FNU
 
 | | PseC aminotransferase with external aldimine | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, aminotransferase | | Authors: | Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
2FN6
 
 | | Helicobacter pylori PseC, aminotransferase involved in the biosynthesis of pseudoaminic acid | | Descriptor: | AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Cygler, M, Lunin, V.V, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structural and Functional Characterization of PseC, an Aminotransferase Involved in the Biosynthesis of Pseudaminic Acid, an Essential Flagellar Modification in Helicobacter pylori
J.Biol.Chem., 281, 2006
|
|
4FMJ
 
 | | Merkel cell polyomavirus VP1 in complex with GD1a oligosaccharide | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
4FMG
 
 | |
4FMH
 
 | | Merkel Cell Polyomavirus VP1 in complex with Disialyllactose | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
4FMI
 
 | | Merkel cell polyomavirus VP1 in complex with 3'-sialyllactosamine | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Hengel, H, Stehle, T. | | Deposit date: | 2012-06-17 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
1XNB
 
 | |
1XND
 
 | |
2C83
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 | | Descriptor: | HYPOTHETICAL PROTEIN PM0188 | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
2C84
 
 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 WITH CMP | | Descriptor: | ALPHA-2,3/2,6-SIALYLTRANSFERASE/SIALIDASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
