3D4S
 
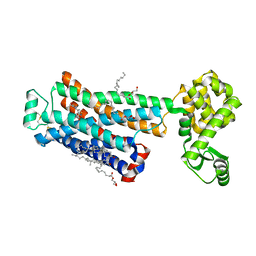 | | Cholesterol bound form of human beta2 adrenergic receptor. | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, Beta-2 adrenergic receptor/T4-lysozyme chimera, ... | | 著者 | Hanson, M.A, Cherezov, V, Roth, C.B, Griffith, M.T, Jaakola, V.-P, Chien, E.Y.T, Velasquez, J, Kuhn, P, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | 登録日 | 2008-05-14 | | 公開日 | 2008-06-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor.
Structure, 16, 2008
|
|
4Z35
 
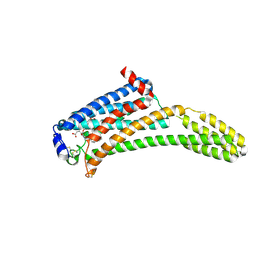 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 | | 分子名称: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 3-{1-[(2S,3S)-3-(4-acetyl-3,5-dimethoxyphenyl)-2-(2,3-dihydro-1H-inden-2-ylmethyl)-3-hydroxypropyl]-4-(methoxycarbonyl)-1H-pyrrol-3-yl}propanoic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | 著者 | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | 登録日 | 2015-03-30 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4Z36
 
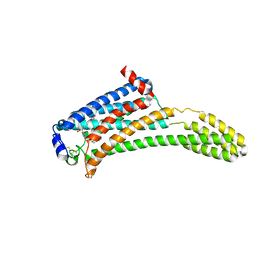 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-3080573 | | 分子名称: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 1-(4-{[(2S,3R)-2-(2,3-dihydro-1H-inden-2-yloxy)-3-(3,5-dimethoxy-4-methylphenyl)-3-hydroxypropyl]oxy}phenyl)cyclopropanecarboxylic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | 著者 | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | 登録日 | 2015-03-30 | | 公開日 | 2015-06-03 | | 最終更新日 | 2015-07-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4Z34
 
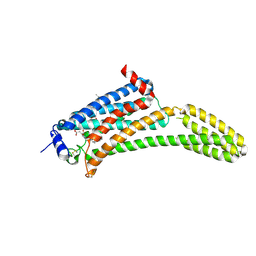 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO9780307 | | 分子名称: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Lysophosphatidic acid receptor 1, Soluble cytochrome b562, ... | | 著者 | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | 登録日 | 2015-03-30 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
6M9T
 
 | | Crystal structure of EP3 receptor bound to misoprostol-FA | | 分子名称: | (11alpha,12alpha,13E,16S)-11,16-dihydroxy-16-methyl-9-oxoprost-13-en-1-oic acid, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | 著者 | Audet, M, White, K.L, Breton, B, Zarzycka, B, Han, G.W, Lu, Y, Gati, C, Batyuk, A, Popov, P, Velasquez, J, Manahan, D, Hu, H, Weierstall, U, Liu, W, Shui, W, Katrich, V, Cherezov, V, Hanson, M.A, Stevens, R.C. | | 登録日 | 2018-08-24 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of misoprostol bound to the labor inducer prostaglandin E2receptor.
Nat. Chem. Biol., 15, 2019
|
|
2FYG
 
 | | Crystal structure of NSP10 from Sars coronavirus | | 分子名称: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | 著者 | Joseph, J.S, Saikatendu, K.S, Subramanian, V, Neuman, B.W, Brooun, A, Griffith, M, Moy, K, Yadav, M.K, Velasquez, J, Buchmeier, M.J, Stevens, R.C, Kuhn, P. | | 登録日 | 2006-02-07 | | 公開日 | 2006-08-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs.
J.Virol., 80, 2006
|
|
1Z9F
 
 | |
1ZX8
 
 | |
2FNO
 
 | |
1J6U
 
 | |
1J5S
 
 | |
2ACF
 
 | | NMR STRUCTURE OF SARS-COV NON-STRUCTURAL PROTEIN NSP3A (SARS1) FROM SARS CORONAVIRUS | | 分子名称: | GLYCEROL, Replicase polyprotein 1ab | | 著者 | Saikatendu, K.S, Joseph, J.S, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2005-07-18 | | 公開日 | 2006-02-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1''-phosphate dephosphorylation by a conserved domain of nsP3.
Structure, 13, 2005
|
|
1VKY
 
 | |
1VKM
 
 | |
1VJO
 
 | |
1VKB
 
 | |
1VLR
 
 | |
1VPZ
 
 | |
1VKH
 
 | |
1VL4
 
 | |
1VRM
 
 | |
1VQ0
 
 | |
1VJ1
 
 | |
1VQR
 
 | |
1VK3
 
 | |
