2F2T
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.7 A resolution with 5-Aminoisoquinoline bound | | Descriptor: | GLYCEROL, ISOQUINOLIN-5-AMINE, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-17 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F8M
 
 | |
2F64
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with 1-METHYLQUINOLIN-2(1H)-ONE bound | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
7TM6
 
 | |
7TMG
 
 | |
7TM7
 
 | |
7TM9
 
 | |
7TM4
 
 | |
7TMB
 
 | |
7TM5
 
 | |
7TM8
 
 | |
7TMD
 
 | |
7TMF
 
 | |
7TMV
 
 | |
7TXX
 
 | |
7UBC
 
 | |
7UBF
 
 | |
7UBD
 
 | |
7UBE
 
 | |
7UCP
 
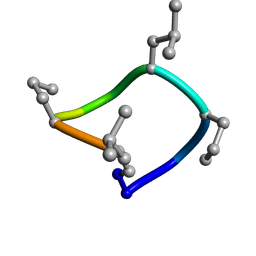 | | computationally designed macrocycle | | Descriptor: | computationally designed cyclic peptide D8.3.p2 | | Authors: | Bhardwaj, G, Baker, D, Rettie, S, Glynn, C, Sawaya, M. | | Deposit date: | 2022-03-17 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Accurate de novo design of membrane-traversing macrocycles.
Cell, 185, 2022
|
|
7UBI
 
 | |
7UBG
 
 | |
7UBH
 
 | |
7UZL
 
 | |
7UWQ
 
 | |
