3T0O
 
 | | Crystal Structure Analysis of Human RNase T2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribonuclease T2 | | Authors: | Thorn, A, Kraetzner, R, Steinfeld, R, Sheldrick, G. | | Deposit date: | 2011-07-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and activity of the only human RNase T2.
Nucleic Acids Res., 40, 2012
|
|
3ST2
 
 | | Dreiklang - equilibrium state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
3ST4
 
 | | Dreiklang - on state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
3TP2
 
 | | Crystal Structure of the Splicing Factor Cwc2 from yeast | | Descriptor: | Pre-mRNA-splicing factor CWC2, SODIUM ION, ZINC ION | | Authors: | Schmitzova, J. | | Deposit date: | 2011-09-07 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Cwc2 reveals a novel architecture of a multipartite RNA-binding protein.
Embo J., 31, 2012
|
|
6ZWF
 
 | | Neisseria gonorrhoeae transaldolase | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, CITRIC ACID, ... | | Authors: | Rabe von Pappenheim, F, Wensien, M, Funk, L.-M, Sautner, V, Tittmann, K. | | Deposit date: | 2020-07-28 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
6ZX4
 
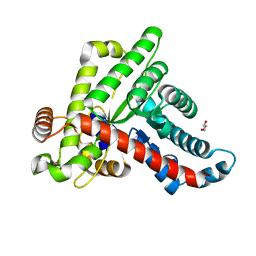 | | Neisseria gonorrhoeae transaldolase | | Descriptor: | CITRIC ACID, GLYCEROL, Transaldolase | | Authors: | Sautner, V, Rabe von Pappenheim, F, Wensien, M, Tittmann, K. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
6ZWJ
 
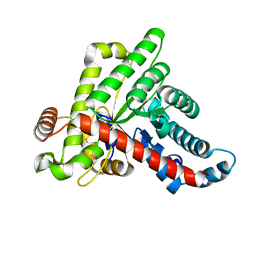 | | Neisseria gonorrhoeae transaldolase at 1.35 Angstrom resolution | | Descriptor: | CITRIC ACID, GLYCEROL, Transaldolase | | Authors: | Rabe von Pappenheim, F, Wensien, M, Sautner, V, Tittmann, K. | | Deposit date: | 2020-07-28 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
6ZWH
 
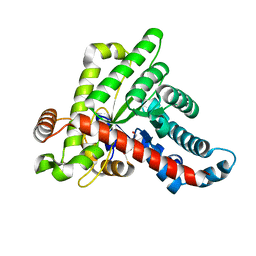 | | Neisseria gonorrhoeae transaldolase at 1.5 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Rabe von Pappenheim, F, Wensien, M, Funk, L.-M, Sautner, V, Tittmann, K. | | Deposit date: | 2020-07-28 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
3C0I
 
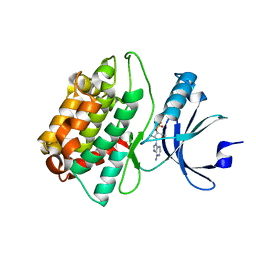 | | CASK CaM-Kinase Domain- 3'-AMP complex, P212121 form | | Descriptor: | Peripheral plasma membrane protein CASK, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Wahl, M.C. | | Deposit date: | 2008-01-20 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | CASK Functions as a Mg2+-independent neurexin kinase
Cell(Cambridge,Mass.), 133, 2008
|
|
3C0H
 
 | | CASK CaM-Kinase Domain- AMPPNP complex, P1 form | | Descriptor: | ADENOSINE MONOPHOSPHATE, Peripheral plasma membrane protein CASK | | Authors: | Wahl, M.C. | | Deposit date: | 2008-01-20 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CASK Functions as a Mg2+-independent neurexin kinase
Cell(Cambridge,Mass.), 133, 2008
|
|
3C0G
 
 | | CASK CaM-Kinase Domain- 3'-AMP complex, P1 form | | Descriptor: | Peripheral plasma membrane protein CASK, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Wahl, M.C. | | Deposit date: | 2008-01-20 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | CASK Functions as a Mg2+-independent neurexin kinase
Cell(Cambridge,Mass.), 133, 2008
|
|
3D3C
 
 | |
3D3B
 
 | |
3FGW
 
 | | One chain form of the 66.3 kDa protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, IODIDE ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
3FGR
 
 | | Two chain form of the 66.3 kDa protein at 1.8 Angstroem | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
3FGT
 
 | | Two chain form of the 66.3 kDa protein from mouse lacking the linker peptide | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
7NQ4
 
 | |
7NZJ
 
 | | Structure of bsTrmB apo | | Descriptor: | GLYCEROL, SODIUM ION, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
7NZI
 
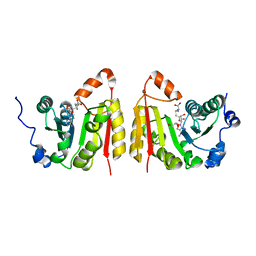 | | TrmB complex with SAH | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
7NYB
 
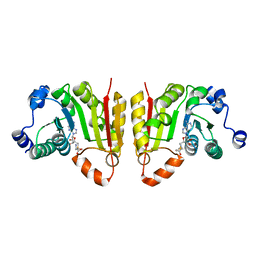 | | TrmB complex with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
7ND2
 
 | | Cryo-EM structure of the human FERRY complex | | Descriptor: | Glutamine amidotransferase-like class 1 domain-containing protein 1, Protein phosphatase 1 regulatory subunit 21, Quinone oxidoreductase-like protein 1 | | Authors: | Quentin, D, Klink, B.U, Raunser, S. | | Deposit date: | 2021-01-29 | | Release date: | 2022-03-02 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mRNA binding by the human FERRY Rab5 effector complex.
Mol.Cell, 83, 2023
|
|
7OOP
 
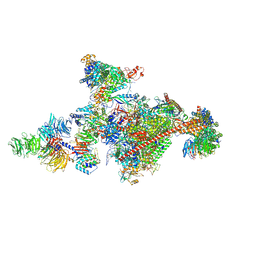 | | Pol II-CSB-CSA-DDB1-UVSSA-PAF-SPT6 (Structure 3) | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OPD
 
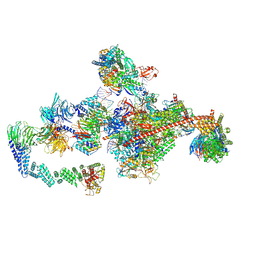 | | Pol II-CSB-CRL4CSA-UVSSA-SPT6-PAF (Structure 5) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OO3
 
 | | Pol II-CSB-CSA-DDB1-UVSSA (Structure1) | | Descriptor: | CSB element, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
7OPC
 
 | | Pol II-CSB-CRL4CSA-UVSSA-SPT6-PAF (Structure 4) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-6, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
