5UAS
 
 | | Structure of a new family of Polysaccharide lyase PL25-Ulvanlyase bound to -[GlcA(1-4)Rha3S]- | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CHLORIDE ION, ... | | Authors: | Ulaganathan, T.S, Cygler, M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Ulvan-Degrading Polysaccharide Lyase Family: Structure and Catalytic Mechanism Suggests Convergent Evolution of Active Site Architecture.
ACS Chem. Biol., 12, 2017
|
|
5UAM
 
 | | Structure of a new family of Polysaccharide lyase PL25-Ulvanlyase. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ulaganathan, T.S, Boniecki, M.T, Cygler, M. | | Deposit date: | 2016-12-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Ulvan-Degrading Polysaccharide Lyase Family: Structure and Catalytic Mechanism Suggests Convergent Evolution of Active Site Architecture.
ACS Chem. Biol., 12, 2017
|
|
5JMD
 
 | | Heparinase III-BT4657 gene product, Methylated Lysines | | Descriptor: | Heparinase III protein, MAGNESIUM ION | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
5JMF
 
 | | Heparinase III-BT4657 gene product | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Heparinase III protein, ... | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
6BYT
 
 | | Complex structure of LOR107 mutant (R320) with tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Ulaganathan, T.S, Cygler, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function analyses of a PL24 family ulvan lyase reveal key features and suggest its catalytic mechanism.
J. Biol. Chem., 293, 2018
|
|
6BYP
 
 | | Structure of PL24 family Polysaccharide lyase-LOR107 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ulaganathan, T.S, Cygler, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analyses of a PL24 family ulvan lyase reveal key features and suggest its catalytic mechanism.
J. Biol. Chem., 293, 2018
|
|
7SNO
 
 | | Structure of Bacple_01701(H214N), a 6-O-galactose porphyran sulfatase | | Descriptor: | 1,2-ETHANEDIOL, 6-O-sulfo-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-6-O-sulfo-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, Arylsulfatase, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
7SNJ
 
 | | Structure of Bacple_01701, a 6-O-galactose porphyran sulfatase | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Arylsulfatase, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
7SNK
 
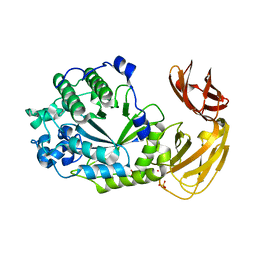 | | Structure of Bacple_01702, a GH29 family glycoside hydrolase | | Descriptor: | Alpha-L-fucosidase, PHOSPHATE ION, POTASSIUM ION | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2021-10-28 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
8EW1
 
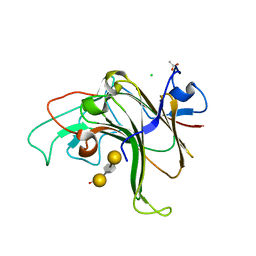 | | Structure of Bacple_01703-E145L | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
8EP4
 
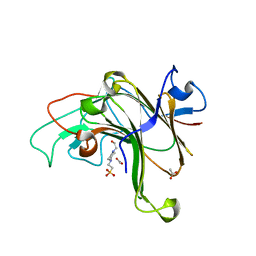 | | Structure of Bacple_01703 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2022-10-05 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
