6G1H
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open form | | 分子名称: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Beloti, L, Frese, A, Mayol, O, Vergne-Vaxelaire, C, Grogan, G. | | 登録日 | 2018-03-21 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
6IAQ
 
 | | Structure of Amine Dehydrogenase from Mycobacterium smegmatis | | 分子名称: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase N-terminus domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Grogan, G, Vaxelaire-Vergne, C, Beloti, L, Mayol, O. | | 登録日 | 2018-11-27 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
6I3G
 
 | | Crystal structure of a putative peptide binding protein AppA from Clostridium difficile | | 分子名称: | ABC transporter, substrate-binding protein, family 5, ... | | 著者 | Hughes, A.M, Wilkinson, A, Dodson, E. | | 登録日 | 2018-11-06 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the putative peptide-binding protein AppA from Clostridium difficile.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8ARN
 
 | | Crystal structure of the peptide binding protein, OppA, from Bacillus subtilis in complex with an endogenous tetrapeptide | | 分子名称: | Endogenous tetrapeptide (SER-ASN-SER-SER), Oligopeptide-binding protein OppA | | 著者 | Hughes, A, Dodson, E.J, Wilkinson, A.J. | | 登録日 | 2022-08-17 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
8AZB
 
 | |
8AY0
 
 | | Crystal Structure of the peptide binding protein DppE from Bacillus subtilis in complex with murein tripeptide | | 分子名称: | 1,2-ETHANEDIOL, Dipeptide-binding protein DppE, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | 著者 | Hughes, A.M, Dodson, E.J, Wilkinson, A.J. | | 登録日 | 2022-09-01 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
8ARE
 
 | | Crystal structure of the peptide binding protein, OppA, from Bacillus subtilis in complex with a PhrE-derived pentapeptide | | 分子名称: | Oligopeptide-binding protein OppA, Phosphatase RapE inhibitor, SULFATE ION | | 著者 | Hughes, A, Dodson, E.J, Wilkinson, A.J. | | 登録日 | 2022-08-16 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
5FWN
 
 | | Imine Reductase from Amycolatopsis orientalis. Closed form in in complex with (R)- Methyltetrahydroisoquinoline | | 分子名称: | (1R)-1-methyl-1,2,3,4-tetrahydroisoquinoline, IMINE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | 登録日 | 2016-02-18 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
8AM3
 
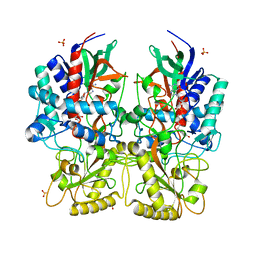 | |
8AM6
 
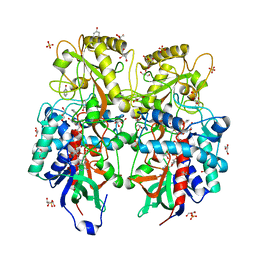 | |
8AM8
 
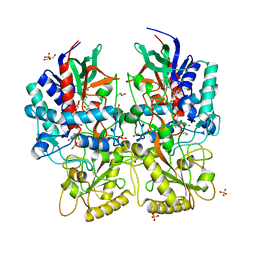 | |
6T9M
 
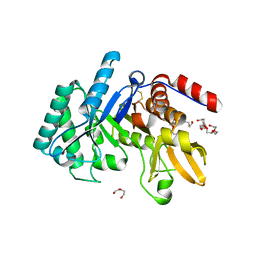 | | Crystal structure of the Chitinase Domain of the Spore Coat Protein CotE from Clostridium difficile | | 分子名称: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptide in active site, ... | | 著者 | Whittingham, J.L, Dodson, E.J, Wilkinson, A.J. | | 登録日 | 2019-10-28 | | 公開日 | 2020-07-22 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structures of the GH18 domain of the bifunctional peroxiredoxin-chitinase CotE from Clostridium difficile.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TSB
 
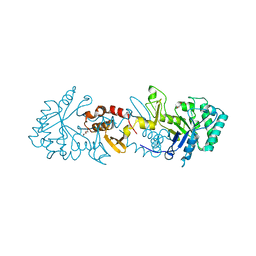 | |
1W0N
 
 | | Structure of uncomplexed Carbohydrate Binding Domain CBM36 | | 分子名称: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE D, MAGNESIUM ION, ... | | 著者 | Jamal, S, Boraston, A.B, Davies, G.J. | | 登録日 | 2004-06-09 | | 公開日 | 2004-10-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (0.8 Å) | | 主引用文献 | Ab Initio Structure Determination and Functional Characterization of Cbm36: A New Family of Calcium-Dependent Carbohydrate Binding Modules
Structure, 12, 2004
|
|
6ZPV
 
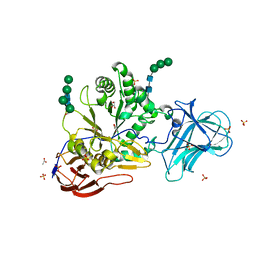 | |
6ZQ0
 
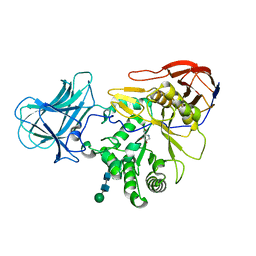 | |
6ZPW
 
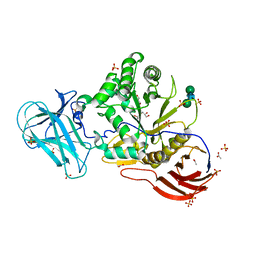 | |
6ZPZ
 
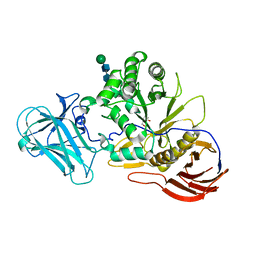 | |
6ZQ1
 
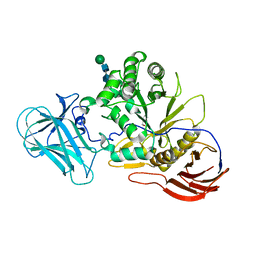 | | Structure of AraDNJ-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | 分子名称: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | McGregor, N.G.S, Davies, G.J. | | 登録日 | 2020-07-09 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPY
 
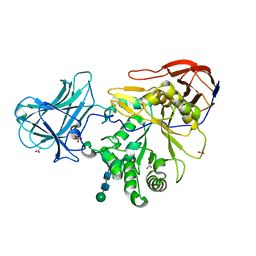 | |
6ZPS
 
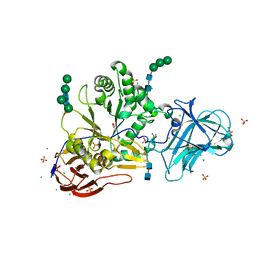 | |
6ZPX
 
 | |
5A9T
 
 | | Imine Reductase from Amycolatopsis orientalis in complex with (R)- Methyltetrahydroisoquinoline | | 分子名称: | (1R)-1-methyl-1,2,3,4-tetrahydroisoquinoline, ACETATE ION, CALCIUM ION, ... | | 著者 | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | 登録日 | 2015-07-22 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
5A9R
 
 | | Apo form of Imine reductase from Amycolatopsis orientalis | | 分子名称: | ACETATE ION, IMINE REDUCTASE | | 著者 | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | 登録日 | 2015-07-22 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
5A9S
 
 | | NADPH complex of Imine Reductase from Amycolatopsis orientalis | | 分子名称: | CALCIUM ION, IMINE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | 登録日 | 2015-07-22 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Stereoselectivity and Structural Characterization of an Imine Reductase (IRED) from Amycolatopsis orientalis
Acs Catalysis, 6, 2016
|
|
