2Y6V
 
 | |
2Y6U
 
 | |
2C1J
 
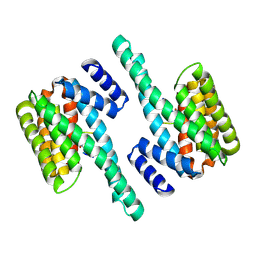 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-15 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
2C1N
 
 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-16 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
3FIL
 
 | |
4CQJ
 
 | |
