1N2D
 
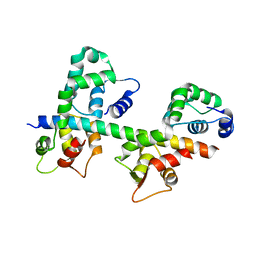 | | Ternary complex of MLC1P bound to IQ2 and IQ3 of Myo2p, a class V myosin | | Descriptor: | IQ2 AND IQ3 MOTIFS FROM MYO2P, A CLASS V MYOSIN, Myosin Light Chain | | Authors: | Terrak, M, Wu, G, Stafford, W.F, Lu, R.C, Dominguez, R. | | Deposit date: | 2002-10-22 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the light chain-binding domain of myosin V.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1M45
 
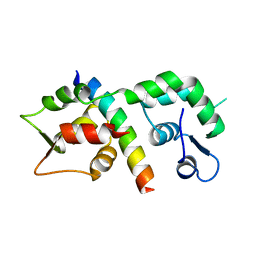 | |
1M46
 
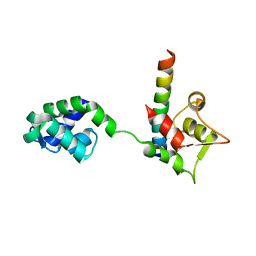 | |
1S70
 
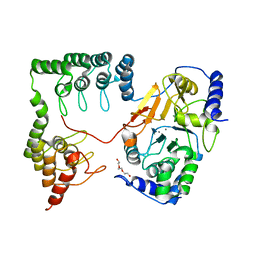 | | Complex between protein ser/thr phosphatase-1 (delta) and the myosin phosphatase targeting subunit 1 (MYPT1) | | Descriptor: | 130 kDa myosin-binding subunit of smooth muscle myosin phophatase (M130), MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-beta (or delta) catalytic subunit, ... | | Authors: | Kerff, F, Terrak, M, Dominguez, R. | | Deposit date: | 2004-01-28 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of protein phosphatase 1 regulation
Nature, 429, 2004
|
|
6ZTG
 
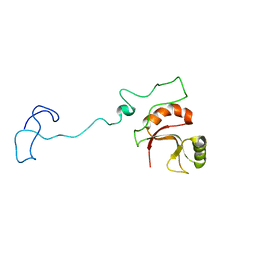 | | Spor protein DedD | | Descriptor: | Cell division protein DedD | | Authors: | Pazos, M, Peters, K, Boes, A, Safaei, Y, Kenward, C, Caveney, N.A, Laguri, C, Breukink, E, Strynadka, N.C.J, Simorre, J.P, Terrak, M, Vollmer, W. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPOR Proteins Are Required for Functionality of Class A Penicillin-Binding Proteins in Escherichia coli.
Mbio, 11, 2020
|
|
6YN0
 
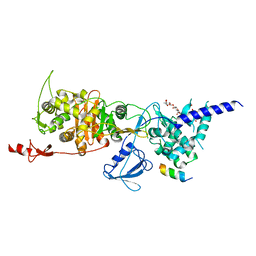 | | Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity | | Descriptor: | Cell division protein FtsN, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Kerff, F, Terrak, M, Boes, A, Herman, H, Charlier, P. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The bacterial cell division protein fragment E FtsN binds to and activates the major peptidoglycan synthase PBP1b.
J.Biol.Chem., 295, 2020
|
|
4BIN
 
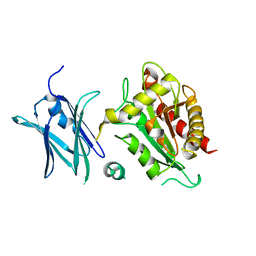 | | Crystal structure of the E. coli N-acetylmuramoyl-L-alanine amidase AmiC | | Descriptor: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE AMIC, SODIUM ION, ZINC ION | | Authors: | Kerff, F, Rocaboy, M, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2013-04-12 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Crystal Structure of the Cell Division Amidase Amic Reveals the Fold of the Amin Domain, a New Peptidoglycan Binding Domain.
Mol.Microbiol., 90, 2013
|
|
