2OSH
 
 | |
3OPX
 
 | |
3ONN
 
 | |
1MC2
 
 | | monomeric LYS-49 phospholipase A2 homologue purified from AG | | Descriptor: | Acutohaemonlysin, ISOPROPYL ALCOHOL | | Authors: | Liu, Q, Huang, Q.Q, Zhang, R.G, Weeks, C.M, Jelsch, C, Teng, M.K, Niu, L.W. | | Deposit date: | 2002-08-05 | | Release date: | 2002-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The crystal structure of a novel, inactive, lysine 49 PLA2 from Agkistrodon acutus venom: an ultrahigh resolution, AB initio structure determination
J.Biol.Chem., 278, 2003
|
|
1XX5
 
 | | Crystal Structure of Natrin from Naja atra snake venom | | Descriptor: | ETHANOL, Natrin 1 | | Authors: | Wang, J, Shen, B, Lou, X.H, Guo, M, Teng, M.K, Niu, L.W. | | Deposit date: | 2004-11-04 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Blocking effect and crystal structure of natrin toxin, a cysteine-rich secretory protein from Naja atra venom that targets the BKCa channel
Biochemistry, 44, 2005
|
|
4ZN3
 
 | | Crystal Structure of MjSpt4:Spt5 complex conformation B | | Descriptor: | FE (III) ION, GLYCEROL, Transcription elongation factor Spt4, ... | | Authors: | Guo, G.R, Zhou, H.H, Gao, Y.X, Zhu, Z.L, Niu, L.W, Teng, M.K. | | Deposit date: | 2015-05-04 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical insights into the DNA-binding mode of MjSpt4p:Spt5 complex at the exit tunnel of RNAPII
J.Struct.Biol., 192, 2015
|
|
4ZN1
 
 | | Crystal Structure of MjSpt4:Spt5 complex conformation A | | Descriptor: | Transcription elongation factor Spt4, Transcription elongation factor Spt5, ZINC ION | | Authors: | Guo, G.R, Zhou, H.H, Gao, Y.X, Zhu, Z.L, Niu, L.W, Teng, M.K. | | Deposit date: | 2015-05-04 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into the DNA-binding mode of MjSpt4p:Spt5 complex at the exit tunnel of RNAPII
J.Struct.Biol., 192, 2015
|
|
5CN2
 
 | |
5CMW
 
 | | Crystal structure of yeast Ent5 N-terminal domain-soaked in KI | | Descriptor: | Epsin-5, GLYCEROL, IODIDE ION | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5CN1
 
 | |
5CMY
 
 | | Crystal structure of yeast Ent5 N-terminal domain-native | | Descriptor: | Epsin-5, GLYCEROL | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Biochem.Biophys.Res.Commun., 477, 2016
|
|
2INF
 
 | | Crystal Structure of Uroporphyrinogen Decarboxylase from Bacillus subtilis | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Fan, J, Liu, Q, Hao, Q, Teng, M.K, Niu, L.W. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of uroporphyrinogen decarboxylase from Bacillus subtilis
J.Bacteriol., 189, 2007
|
|
3KW7
 
 | | Crystal structure of LacB from Trametes sp. AH28-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Ge, H.H, Teng, M.K, Niu, L.W. | | Deposit date: | 2009-12-01 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Structure of native laccase B from Trametes sp. AH28-2
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2HDZ
 
 | | Crystal Structure Analysis of the UBF HMG box5 | | Descriptor: | Nucleolar transcription factor 1 | | Authors: | Rong, H, Teng, M.K, Niu, L.W. | | Deposit date: | 2006-06-21 | | Release date: | 2007-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human upstream binding factor HMG box 5 and site for binding of the cell-cycle regulatory factor TAF1
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5HS5
 
 | |
5J08
 
 | |
3EWG
 
 | | Crystal structure of the N-terminal domain of NusG (NGN) from Methanocaldococcus jannaschii | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Putative transcription antitermination protein nusG | | Authors: | Zhou, H.H, Liu, Q, Gao, Y.X, Teng, M.K, Niu, L.W. | | Deposit date: | 2008-10-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of NusG N-terminal (NGN) domain from Methanocaldococcus jannaschii and its interaction with rpoE''
Proteins, 76, 2009
|
|
3U53
 
 | | Crystal structure of human Ap4A hydrolase | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase [asymmetrical], GLYCEROL, SULFATE ION | | Authors: | Ge, H.H, Teng, M.K. | | Deposit date: | 2011-10-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Crystal structure of wild-type and mutant human Ap4A hydrolase
Biochem.Biophys.Res.Commun., 432, 2013
|
|
3TO8
 
 | | Crystal structure of the two C-terminal RRM domains of heterogeneous nuclear ribonucleoprotein L (hnRNP L) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Heterogeneous nuclear ribonucleoprotein L, ... | | Authors: | Zhang, W.J, Zeng, F.X, Liu, Y.W, Zhao, Y, Niu, L.W, Teng, M.K, Li, X. | | Deposit date: | 2011-09-04 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of the two C-terminal RRM domains of heterogeneous nuclear ribonucleoprotein L (hnRNP L)
To be Published
|
|
5YL6
 
 | | Crystal structure of Ofd2 in complex with 2OG from Schizosaccharomyces pombe | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | Authors: | Wang, J, Gao, Y.X, Guo, G.R, Zhu, Z.L, Teng, M.K, Niu, L.W. | | Deposit date: | 2017-10-17 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Ofd2 in complex with 2OG from Schizosaccharomyces pombe
to be published
|
|
5YLB
 
 | | Crystal structure of Ofd2 from Schizosaccharomyces pombe at 1.80 A | | Descriptor: | FE (III) ION, Uncharacterized protein P8A3.02c | | Authors: | Wang, J, Gao, Y.X, Guo, G.R, Zhu, Z.L, Teng, M.K, Niu, L.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of Ofd2 from Schizosaccharomyces pombe at 1.80 A
to be published
|
|
4HBG
 
 | |
4IFS
 
 | | Crystal structure of the hSSRP1 Middle domain | | Descriptor: | CHLORIDE ION, FACT complex subunit SSRP1 | | Authors: | Zhang, W.J, Zeng, F.X, Shao, C, Liu, Y.W, Niu, L.W, Li, X, Teng, M.K. | | Deposit date: | 2012-12-15 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the hSSRP1 Middle domain
To be Published
|
|
1DNH
 
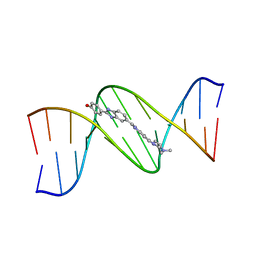 | | THE MOLECULAR STRUCTURE OF THE COMPLEX OF HOECHST 33258 AND THE DNA DODECAMER D(CGCGAATTCGCG) | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Teng, M.-K, Usman, N, Frederick, C.A, Wang, A.H.-J. | | Deposit date: | 1988-02-16 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG).
Nucleic Acids Res., 16, 1988
|
|
4YLY
 
 | | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, staphylococcus aureus at 2.25 angstrom resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-03-06 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Staphylococcus aureus peptidyl-tRNA hydrolase at a 2.25 angstrom resolution.
Acta Biochim.Biophys.Sin., 47, 2015
|
|
