4MS2
 
 | | Structural basis of Ca2+ selectivity of a voltage-gated calcium channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MTF
 
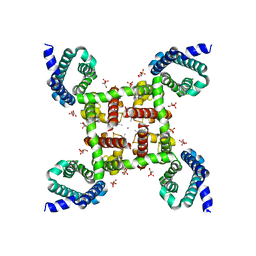 | | Structural Basis of Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MTG
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MVM
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1997 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MVU
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MW8
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.256 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MVO
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
8H8A
 
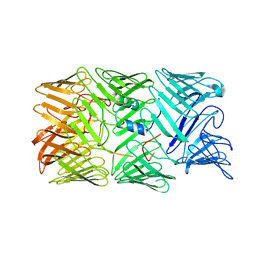 | | Type VI secretion system effector RhsP in its post-autoproteolysis and monomeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
8H8C
 
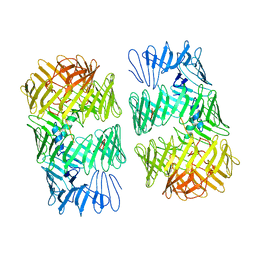 | | Type VI secretion system effector RhsP in its post-autoproteolysis and dimeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
8H8B
 
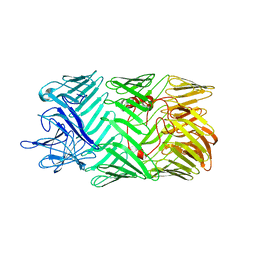 | | Type VI secretion system effector RhsP in its pre-autoproteolysis and monomeric form | | Descriptor: | Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
5VGT
 
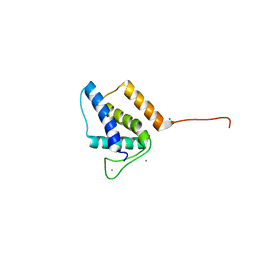 | | X-ray structure of bacteriophage Sf6 tail adaptor protein gp7 | | Descriptor: | CALCIUM ION, Gene 7 protein, MAGNESIUM ION | | Authors: | Tang, L, Liang, L, Zhao, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | High-resolution structure of podovirus tail adaptor suggests repositioning of an octad motif that mediates the sequential tail assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6JUH
 
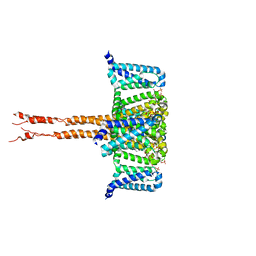 | | structure of CavAb in complex with efonidipine | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[phenyl-(phenylmethyl)amino]ethyl (4~{R})-5-(5,5-dimethyl-2-oxidanylidene-1,3,2$l^{5}-dioxaphosphinan-2-yl)-2,6-dimethyl-4-(3-nitrophenyl)-1,4-dihydropyridine-3-carboxylate, ... | | Authors: | Tang, L, Xu, F. | | Deposit date: | 2019-04-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for efonidipine block of a voltage-gated Ca2+channel.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
1I0E
 
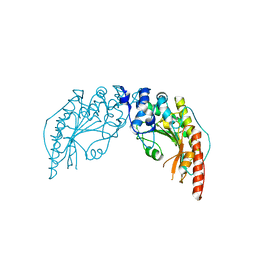 | | CRYSTAL STRUCTURE OF CREATINE KINASE FROM HUMAN MUSCLE | | Descriptor: | CREATINE KINASE,M CHAIN | | Authors: | Shen, Y.-Q, Tang, L, Zhou, H.-M, Lin, Z.-J. | | Deposit date: | 2001-01-29 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of human muscle creatine kinase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3WRO
 
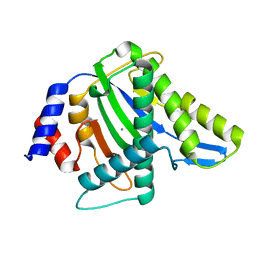 | |
3WRN
 
 | |
3WRS
 
 | |
3WRR
 
 | |
3WRQ
 
 | |
1BK9
 
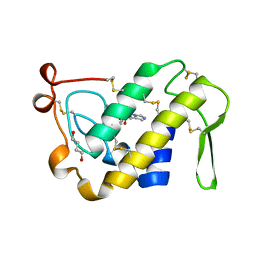 | | PHOSPHOLIPASE A2 MODIFIED BY PBPB | | Descriptor: | 1,4-BUTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2, ... | | Authors: | Zhao, H, Tang, L, Wang, X, Lin, Z, Zhou, Y. | | Deposit date: | 1998-07-16 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a snake venom phospholipase A2 modified by p-bromo-phenacyl-bromide.
Toxicon, 36, 1998
|
|
1D9K
 
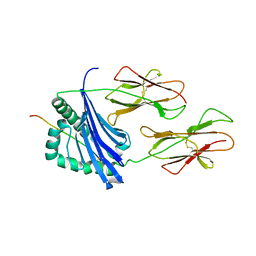 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | Authors: | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | Deposit date: | 1999-10-28 | | Release date: | 1999-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
3ZN2
 
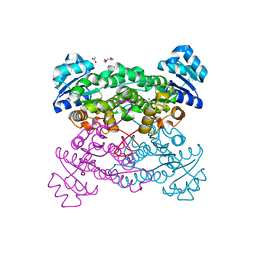 | | protein engineering of halohydrin dehalogenase | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETATE ION, HALOHYDRIN DEHALOGENASE, ... | | Authors: | Schallmey, M, Jekel, P, Tang, L, Majeric-Elenkov, M, Hoeffken, H.W, Hauer, B, Janssen, D.B. | | Deposit date: | 2013-02-13 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Single Point Mutation Enhances Hydroxynitrile Synthesis by Halohydrin Dehalogenase.
Enzyme.Microb.Technol., 70, 2015
|
|
2F7K
 
 | |
3NB3
 
 | | The host outer membrane proteins OmpA and OmpC are packed at specific sites in the Shigella phage Sf6 virion as structural components | | Descriptor: | Outer membrane protein A, Outer membrane protein C | | Authors: | Zhao, H, Sequeira, R.D, Galeva, N.A, Tang, L. | | Deposit date: | 2010-06-02 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | The host outer membrane proteins OmpA and OmpC are associated with the Shigella phage Sf6 virion.
Virology, 409, 2011
|
|
3EOP
 
 | | Crystal Structure of the DUF55 domain of human thymocyte nuclear protein 1 | | Descriptor: | SULFATE ION, Thymocyte nuclear protein 1 | | Authors: | Yu, F, Song, A, Xu, C, Sun, L, Li, L, Tang, L, Hu, H, He, J. | | Deposit date: | 2008-09-29 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determining the DUF55-domain structure of human thymocyte nuclear protein 1 from crystals partially twinned by tetartohedry
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5L35
 
 | |
