4FK9
 
 | |
4TYV
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4TZ1
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose | | Descriptor: | Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4TZ5
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4TZ3
 
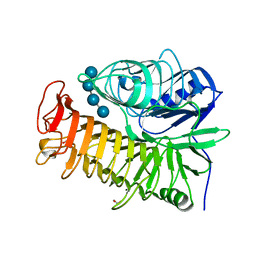 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4IM4
 
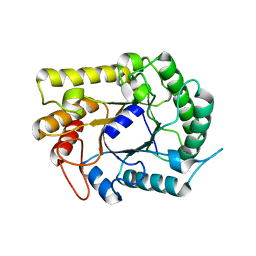 | |
4ILT
 
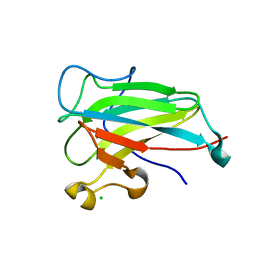 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | CHLORIDE ION, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2012-12-31 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
4ILV
 
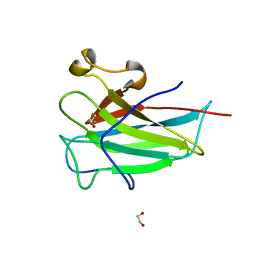 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
4PEX
 
 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4PEW
 
 | | Structure of sacteLam55A from Streptomyces sp. SirexAA-E | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Putative secreted protein | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4PEY
 
 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose | | Descriptor: | Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4PEZ
 
 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4PF0
 
 | | Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-04-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
6UI3
 
 | | GH5-4 broad specificity endoglucanase from Clostridum cellulovorans | | Descriptor: | 1,2-ETHANEDIOL, Cellulase | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6MQ4
 
 | | GH5-4 broad specificity endoglucanase from Hungateiclostridium cellulolyticum | | Descriptor: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2018-10-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6Q1I
 
 | |
6PZ7
 
 | | GH5-4 broad specificity endoglucanase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Endoglucanase family 5 | | Authors: | Bianchetti, C.M, Bingman, C.A, Fox, B.G. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6WQV
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose | | Descriptor: | 1,2-ETHANEDIOL, Endoglucanase, NITRATE ION, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6WQY
 
 | | GH5-4 broad specificity endoglucanase from Bacteroides salanitronis | | Descriptor: | CHLORIDE ION, Cellulase, MAGNESIUM ION | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6WQP
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Endoglucanase, ... | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6XSU
 
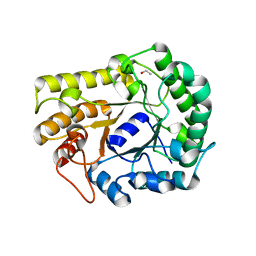 | | GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens | | Descriptor: | ETHANOL, GH5-4 broad specificity endoglucanase | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6XRK
 
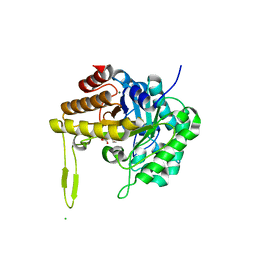 | | GH5-4 broad specificity endoglucanase from an uncultured bovine rumen ciliate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-13 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6XSO
 
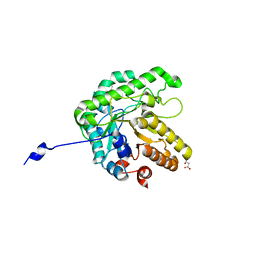 | | GH5-4 broad specificity endoglucanase from an uncultured bacterium | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Cellulase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-07-15 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
4ILY
 
 | |
