1FUW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A DOUBLE MUTANT SINGLE-CHAIN MONELLIN(SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | MONELLIN | | Authors: | Sung, Y.H, Shin, J, Jung, J, Lee, W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics, and stability of a double mutant single-chain monellin. structural origin of sweetness.
J.Biol.Chem., 276, 2001
|
|
8Z0R
 
 | | Cryo-EM structure of tetramer HtmB2-CT | | Descriptor: | special condensation domain in NRPS | | Authors: | Sun, Y.H, Zhang, Z.Y, Mei, Q. | | Deposit date: | 2024-04-10 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Formation of the Diketopiperazine Moiety by a Distinct Condensation-Like Domain in Hangtaimycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 2025
|
|
8Z0S
 
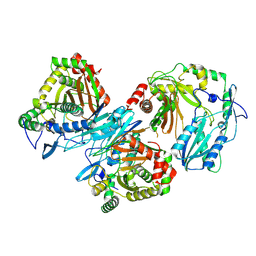 | | Cryo-EM structure of trimer HtmB2-CT | | Descriptor: | special condensation domain in NRPS | | Authors: | Sun, Y.H, Zhang, Z.Y, Mei, Q. | | Deposit date: | 2024-04-10 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Formation of the Diketopiperazine Moiety by a Distinct Condensation-Like Domain in Hangtaimycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 2025
|
|
8Z0Q
 
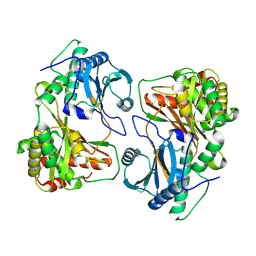 | | Cryo-EM structure of dimer HtmB2-CT | | Descriptor: | Special condensation domain in NRPS | | Authors: | Sun, Y.H, Zhang, Z.Y, Mei, Q. | | Deposit date: | 2024-04-10 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Formation of the Diketopiperazine Moiety by a Distinct Condensation-Like Domain in Hangtaimycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 2025
|
|
5YDM
 
 | |
9C45
 
 | |
9C44
 
 | |
9C6O
 
 | |
9DAK
 
 | |
5YDL
 
 | |
5YDA
 
 | |
8ZUF
 
 | | Cryo-EM structure of P.nat ACE2 mutant in complex with MOW15-22 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Tang, J, Deng, Z. | | Deposit date: | 2024-06-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses.
Cell, 188, 2025
|
|
