5Z0I
 
 | |
5Z0H
 
 | |
5Z0G
 
 | |
3AZO
 
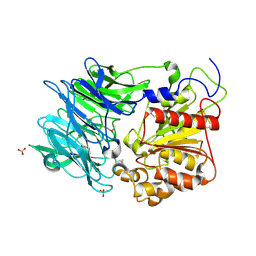 | | Crystal structure of puromycin hydrolase | | 分子名称: | Aminopeptidase, SULFATE ION | | 著者 | Matoba, Y, Sugiyama, M. | | 登録日 | 2011-05-27 | | 公開日 | 2011-07-27 | | 最終更新日 | 2011-09-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural evidence that puromycin hydrolase is a new type of aminopeptidase with a prolyl oligopeptidase family fold
Proteins, 79, 2011
|
|
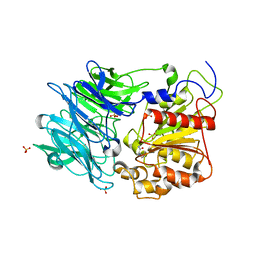
3AZP
 
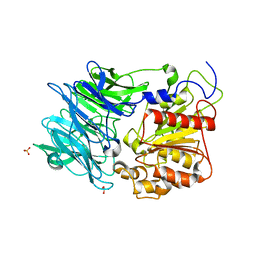 | |
3AZQ
 
 | |
3WSV
 
 | |
3WSW
 
 | |
2ZRR
 
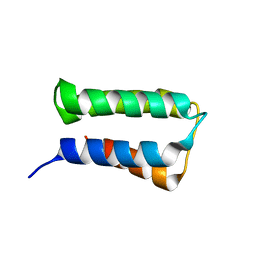 | | Crystal structure of an immunity protein that contributes to the self-protection of bacteriocin-producing Enterococcus mundtii 15-1A | | 分子名称: | Mundticin KS immunity protein | | 著者 | Jeon, H.J, Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | 登録日 | 2008-08-30 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure and mutagenic analysis of a bacteriocin immunity protein, Mun-im
Biochem.Biophys.Res.Commun., 378, 2009
|
|
3VVL
 
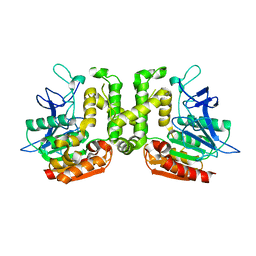 | | Crystal structure of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | 分子名称: | Homoserine O-acetyltransferase | | 著者 | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | 登録日 | 2012-07-26 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
3VVM
 
 | | Crystal structure of G52A-P55G mutant of L-serine-O-acetyltransferase found in D-cycloserine biosynthetic pathway | | 分子名称: | Homoserine O-acetyltransferase | | 著者 | Oda, K, Matoba, Y, Kumagai, T, Noda, M, Sugiyama, M. | | 登録日 | 2012-07-26 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystallographic study to determine the substrate specificity of an L-serine-acetylating enzyme found in the D-cycloserine biosynthetic pathway
J.Bacteriol., 195, 2013
|
|
2ZW7
 
 | |
2ZW5
 
 | |
2ZW6
 
 | |
2ZW4
 
 | |
3WA6
 
 | |
3WA7
 
 | | Crystal structure of selenomethionine-labeled tannase from Lactobacillus plantarum in the orthorhombic crystal | | 分子名称: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Matoba, Y, Tanaka, N, Sugiyama, M. | | 登録日 | 2013-04-27 | | 公開日 | 2013-07-24 | | 最終更新日 | 2013-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystallographic and mutational analyses of tannase from Lactobacillus plantarum.
Proteins, 81, 2013
|
|
2ZV6
 
 | | Crystal structure of human squamous cell carcinoma antigen 1 | | 分子名称: | Serpin B3 | | 著者 | Zheng, B, Matoba, Y, Katagiri, C, Hibino, T, Sugiyama, M. | | 登録日 | 2008-11-01 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of SCCA1 and insight about the interaction with JNK1
Biochem.Biophys.Res.Commun., 380, 2009
|
|
3X44
 
 | | Crystal structure of O-ureido-L-serine-bound K43A mutant of O-ureido-L-serine synthase | | 分子名称: | (E)-O-(carbamoylamino)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, O-ureido-L-serine synthase | | 著者 | Matoba, Y, Uda, N, Oda, K, Sugiyama, M. | | 登録日 | 2015-03-13 | | 公開日 | 2015-07-29 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structural and mutational analyses of O-ureido-L-serine synthase necessary for D-cycloserine biosynthesis.
Febs J., 282, 2015
|
|
3X43
 
 | | Crystal structure of O-ureido-L-serine synthase | | 分子名称: | O-ureido-L-serine synthase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matoba, Y, Uda, N, Oda, K, Sugiyama, M. | | 登録日 | 2015-03-13 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The structural and mutational analyses of O-ureido-L-serine synthase necessary for D-cycloserine biosynthesis.
Febs J., 282, 2015
|
|
8ILL
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH5.6 | | 分子名称: | CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, green fluorescent protein | | 著者 | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | 登録日 | 2023-03-03 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8ILK
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH8.5 | | 分子名称: | CHLORIDE ION, Green FLUORESCENT PROTEIN | | 著者 | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | 登録日 | 2023-03-03 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8IHL
 
 | | Overlapping tri-nucleosome | | 分子名称: | DNA (353-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Nishimura, M, Fujii, T, Tanaka, H, Maehara, K, Nozawa, K, Takizawa, Y, Ohkawa, Y, Kurumizaka, H. | | 登録日 | 2023-02-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (7.64 Å) | | 主引用文献 | Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties.
Commun Biol, 7, 2024
|
|
6V2K
 
 | | The nucleosome structure after H2A-H2B exchange | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | 著者 | Arimura, Y, Hirano, R, Kurumizaka, H. | | 登録日 | 2019-11-24 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A-H2B in the nucleosome.
Commun Biol, 4, 2021
|
|
5GSE
 
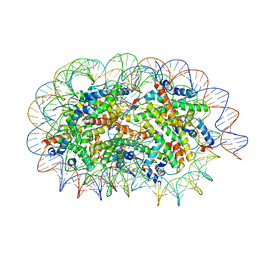 | | Crystal structure of unusual nucleosome | | 分子名称: | DNA (250-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Kato, D, Osakabe, A, Arimura, Y, Park, S.Y, Kurumizaka, H. | | 登録日 | 2016-08-16 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Crystal structure of the overlapping dinucleosome composed of hexasome and octasome
Science, 356, 2017
|
|
