2RSG
 
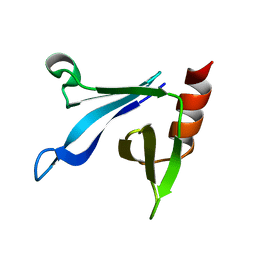 | | Solution structure of the CERT PH domain | | 分子名称: | Collagen type IV alpha-3-binding protein | | 著者 | Sugiki, T, Takeuchi, K, Tokunaga, Y, Kumagai, K, Kawano, M, Nishijima, M, Hanada, K, Takahashi, H, Shimada, I. | | 登録日 | 2012-02-25 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the Golgi association by the pleckstrin homology domain of the ceramide trafficking protein (CERT)
J.Biol.Chem., 287, 2012
|
|
5GIW
 
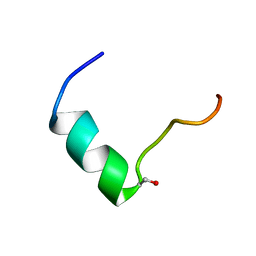 | | Solution NMR structure of Humanin containing a D-isomerized serine residue | | 分子名称: | Humanin | | 著者 | Furuita, K, Sugiki, T, Alsanousi, N, Fujiwara, T, Kojima, C. | | 登録日 | 2016-06-25 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure and inhibitory effect against amyloid-beta fibrillation of Humanin containing a d-isomerized serine residue
Biochem.Biophys.Res.Commun., 477, 2016
|
|
6LLQ
 
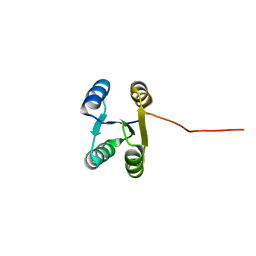 | | Solution NMR structure of de novo Rossmann2x2 fold with most of the core mutated to valine, R2x2_VAL88 | | 分子名称: | VAL88 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Koga, R, Yamamoto, M, Kosugi, T, Koga, N. | | 登録日 | 2019-12-23 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Robust folding of a de novo designed ideal protein even with most of the core mutated to valine.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2ND5
 
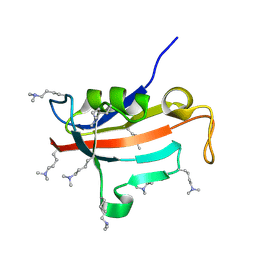 | | Lysine dimethylated FKBP12 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1A | | 著者 | Hattori, Y, Sebera, J, Sychrovsky, V, Furuita, K, Sugiki, T, Ohki, I, Ikegami, T, Kobayashi, N, Tanaka, Y, Fujiwara, T, Kojima, C. | | 登録日 | 2016-05-05 | | 公開日 | 2017-05-17 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Observation of Protein Surface Salt Bridges at Neutral pH
To be Published
|
|
2RUJ
 
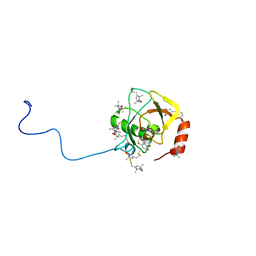 | | Solution structure of MTSL spin-labeled Schizosaccharomyces pombe Sin1 CRIM domain | | 分子名称: | Stress-activated map kinase-interacting protein 1 | | 著者 | Furuita, K, Kataoka, S, Sugiki, T, Kobayashi, N, Ikegami, T, Shiozaki, K, Fujiwara, T, Kojima, C. | | 登録日 | 2014-07-24 | | 公開日 | 2015-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Utilization of paramagnetic relaxation enhancements for high-resolution NMR structure determination of a soluble loop-rich protein with sparse NOE distance restraints
J.Biomol.Nmr, 61, 2015
|
|
7BPP
 
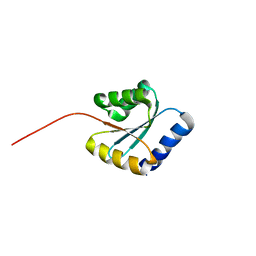 | | Solution NMR structure of NF5; de novo designed protein with a novel fold | | 分子名称: | NF5 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-23 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-07-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPM
 
 | | Solution NMR structure of NF2; de novo designed protein with a novel fold | | 分子名称: | NF2 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-23 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-07-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQD
 
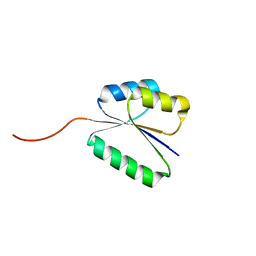 | | Solution NMR structure of NF8 (knot fold); de novo designed protein with a novel fold | | 分子名称: | NF8 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-07-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPN
 
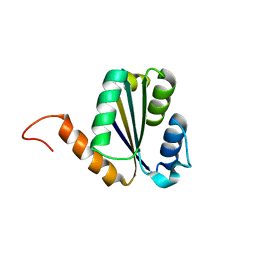 | | Solution NMR structure of NF7; de novo designed protein with a novel fold | | 分子名称: | NF7 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-23 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-07-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPL
 
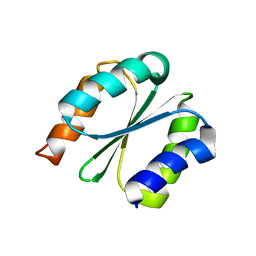 | | Solution NMR structure of NF1; de novo designed protein with a novel fold | | 分子名称: | NF1 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-23 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-07-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQQ
 
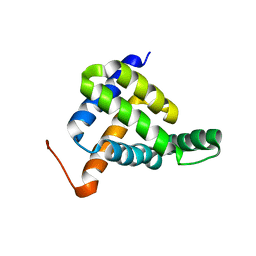 | | Solution NMR structure of fold-Z Gogy; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Gogy | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQM
 
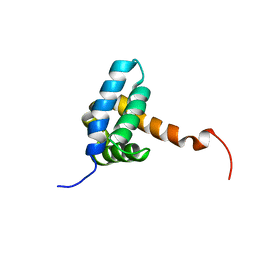 | | Solution NMR structure of fold-0 Chantal; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Chantal | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQN
 
 | | Solution NMR structure of fold-C Rei; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Rei | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQE
 
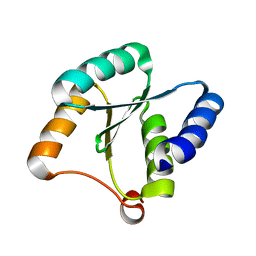 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | 分子名称: | NF3 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-07-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
4U5X
 
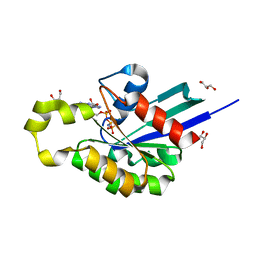 | | Structure of plant small GTPase OsRac1 complexed with the non-hydrolyzable GTP analog GMPPNP | | 分子名称: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Ohki, I, Kosami, K, Fujiwara, T, Nakagawa, A, Shimamoto, K, Kojima, C. | | 登録日 | 2014-07-25 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Crystal Structure of the Plant Small GTPase OsRac1 Reveals Its Mode of Binding to NADPH Oxidase
J.Biol.Chem., 289, 2014
|
|
7BQS
 
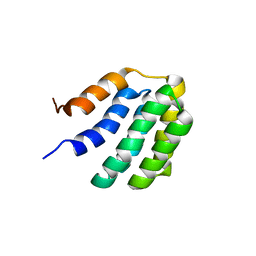 | | Solution NMR structure of fold-U Nomur; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Nomur | | 著者 | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQR
 
 | | Solution NMR structure of fold-K Mussoc; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Mussoc | | 著者 | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7DNS
 
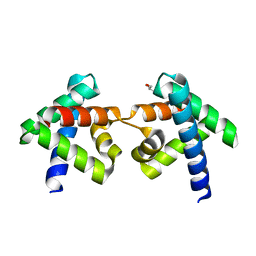 | | Crystal structure of domain-swapped dimer of H5_Fold-0 Elsa; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | GLYCEROL, de novo designed protein | | 著者 | Suzuki, K, Kobayashi, N, Murata, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-12-10 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.327 Å) | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQC
 
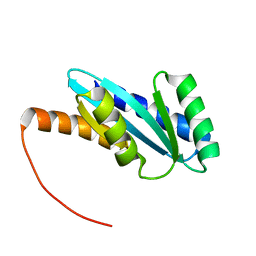 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | 分子名称: | NF4 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-07-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQB
 
 | | Solution NMR structure of NF6; de novo designed protein with a novel fold | | 分子名称: | NF6 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-07-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
