2DNJ
 
 | |
3DNI
 
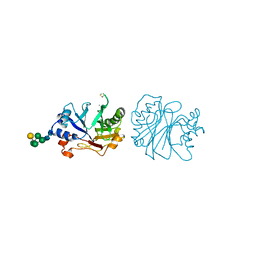 | | CRYSTALLOGRAPHIC REFINEMENT AND STRUCTURE OF DNASE I AT 2 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, DEOXYRIBONUCLEASE I, alpha-D-galactopyranose-(1-6)-beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oefner, C, Suck, D. | | Deposit date: | 1992-08-20 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic refinement and structure of DNase I at 2 A resolution.
J.Mol.Biol., 192, 1986
|
|
1ATN
 
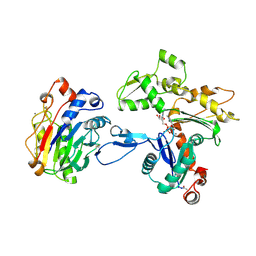 | | Atomic structure of the actin:DNASE I complex | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kabsch, W, Mannherz, H.G, Suck, D, Pai, E, Holmes, K.C. | | Deposit date: | 1991-03-08 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the actin:DNase I complex.
Nature, 347, 1990
|
|
2QNC
 
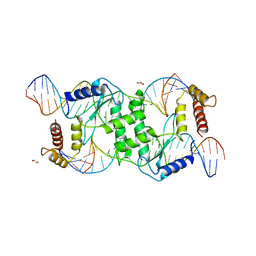 | | Crystal structure of T4 Endonuclease VII N62D mutant in complex with a DNA Holliday junction | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DGP*DGP*DCP*DCP*DTP*DAP*DGP*DCP*DGP*DTP*DCP*DCP*DGP*DGP*DAP*DAP*DTP*DTP*DCP*DTP*DTP*DCP*DG)-3'), DNA (5'-D(*DCP*DAP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DGP*DAP*DGP*DCP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DCP*DCP*DT)-3'), ... | | Authors: | Biertumpfel, C, Yang, W, Suck, D. | | Deposit date: | 2007-07-18 | | Release date: | 2008-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of T4 endonuclease VII resolving a Holliday junction.
Nature, 449, 2007
|
|
1LJO
 
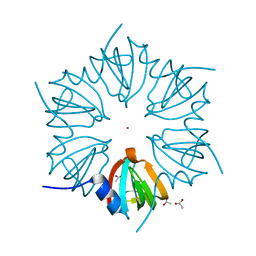 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM2) FROM ARCHAEOGLOBUS FULGIDUS AT 1.95A RESOLUTION | | Descriptor: | ACETIC ACID, Archaeal Sm-like protein AF-Sm2, CADMIUM ION | | Authors: | Toro, I, Basquin, J, Teo-Dreher, H, Suck, D. | | Deposit date: | 2002-04-22 | | Release date: | 2002-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Archaeal Sm proteins form heptameric and hexameric complexes: crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus.
J.Mol.Biol., 320, 2002
|
|
1DNK
 
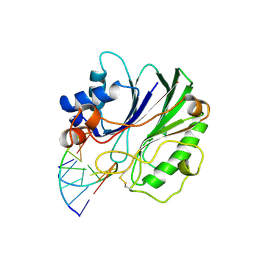 | | THE X-RAY STRUCTURE OF THE DNASE I-D(GGTATACC)2 COMPLEX AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*GP*GP*TP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3'), ... | | Authors: | Weston, S.A, Lahm, A, Suck, D. | | Deposit date: | 1992-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution.
J.Mol.Biol., 226, 1992
|
|
1AK0
 
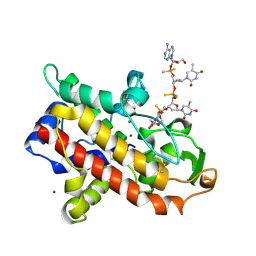 | | P1 NUCLEASE IN COMPLEX WITH A SUBSTRATE ANALOG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-(DITHIO)PHOSPHATE, ... | | Authors: | Romier, C, Suck, D. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of single-stranded DNA by nuclease P1: high resolution crystal structures of complexes with substrate analogs.
Proteins, 32, 1998
|
|
1LFB
 
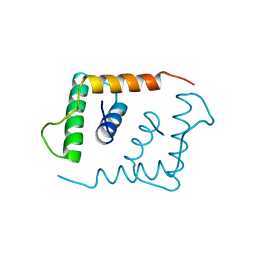 | | THE X-RAY STRUCTURE OF AN ATYPICAL HOMEODOMAIN PRESENT IN THE RAT LIVER TRANSCRIPTION FACTOR LFB1(SLASH)HNF1 AND IMPLICATIONS FOR DNA BINDING | | Descriptor: | LIVER TRANSCRIPTION FACTOR (LFB1) | | Authors: | Ceska, T.A, Lamers, M, Monaci, P, Nicosia, A, Cortese, R, Suck, D. | | Deposit date: | 1993-06-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of an atypical homeodomain present in the rat liver transcription factor LFB1/HNF1 and implications for DNA binding.
EMBO J., 12, 1993
|
|
1M8V
 
 | | Structure of Pyrococcus abyssii Sm Protein in Complex with a Uridine Heptamer | | Descriptor: | 5'-R(P*UP*UP*UP*UP*UP*UP*U)-3', CALCIUM ION, PUTATIVE SNRNP SM-LIKE PROTEIN, ... | | Authors: | Thore, S, Mayer, C, Sauter, C, Weeks, S, Suck, D. | | Deposit date: | 2002-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Pyrococcus abyssii Sm core and its Complex with RNA: Common Features of RNA-binding in Archaea and Eukarya
J.Biol.Chem., 278, 2003
|
|
1WKD
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1WKE
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
1WKF
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-08-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile.
Biochemistry, 35, 1996
|
|
2HQT
 
 | |
1PUD
 
 | | TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Romier, C, Reuter, K, Suck, D, Ficner, R. | | Deposit date: | 1996-06-28 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange.
EMBO J., 15, 1996
|
|
2HRA
 
 | | Crystal structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA aminoacylation and nuclear export cofactor Arc1p reveal a novel function for an old fold | | Descriptor: | Glutamyl-tRNA synthetase, cytoplasmic, IODIDE ION | | Authors: | Simader, H, Hothorn, M, Suck, D. | | Deposit date: | 2006-07-20 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA-aminoacylation and nuclear-export cofactor Arc1p reveal a novel function for an old fold.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1XO1
 
 | | T5 5'-EXONUCLEASE MUTANT K83A | | Descriptor: | 5'-EXONUCLEASE | | Authors: | Ceska, T.A, Suck, D, Sayers, J.R. | | Deposit date: | 1998-11-19 | | Release date: | 1999-04-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis of conserved lysine residues in bacteriophage T5 5'-3' exonuclease suggests separate mechanisms of endo-and exonucleolytic cleavage.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1E7L
 
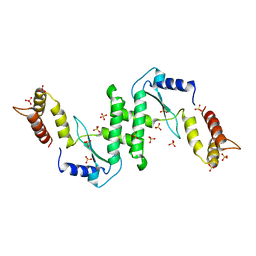 | | Endonuclease VII (EndoVII) N62D mutant from phage T4 | | Descriptor: | RECOMBINATION ENDONUCLEASE VII, SULFATE ION, ZINC ION | | Authors: | Raaijmakers, H.C.A, Vix, O, Toro, I, Suck, D. | | Deposit date: | 2000-08-29 | | Release date: | 2001-05-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Conformational Flexibility in T4 Endonuclease Vii Revealed by Crystallography: Implications for Substrate Binding and Cleavage
J.Mol.Biol., 308, 2001
|
|
1EXN
 
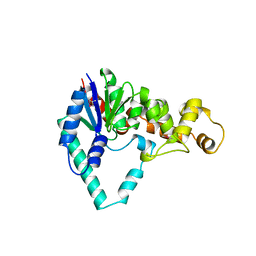 | | T5 5'-EXONUCLEASE | | Descriptor: | 5'-EXONUCLEASE | | Authors: | Ceska, T.A, Sayers, J.R, Stier, G, Suck, D. | | Deposit date: | 1997-01-17 | | Release date: | 1997-07-07 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A helical arch allowing single-stranded DNA to thread through T5 5'-exonuclease.
Nature, 382, 1996
|
|
1E7D
 
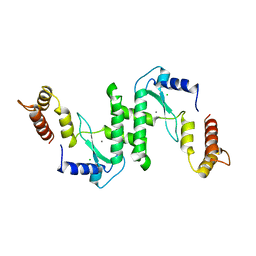 | | Endonuclease VII (ENDOVII) Ffrom Phage T4 | | Descriptor: | CALCIUM ION, CHLORIDE ION, RECOMBINATION ENDONUCLEASE VII, ... | | Authors: | Raaijmakers, H.C.A, Vix, O, Toro, I, Suck, D. | | Deposit date: | 2000-08-28 | | Release date: | 2001-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Flexibility in T4 Endonuclease Vii Revealed by Crystallography: Implications for Substrate Binding and Cleavage
J.Mol.Biol., 308, 2001
|
|
1UT5
 
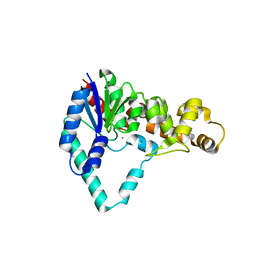 | |
1UT8
 
 | |
1I5L
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS COMPLEXED WITH SHORT POLY-U RNA | | Descriptor: | 5'-R(*UP*UP*U)-3', PUTATIVE SNRNP SM-LIKE PROTEIN AF-SM1, URIDINE | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-28 | | Release date: | 2001-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
1HK9
 
 | |
1I4K
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS AT 2.5A RESOLUTION | | Descriptor: | CITRIC ACID, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-22 | | Release date: | 2001-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
1HLO
 
 | | THE CRYSTAL STRUCTURE OF AN INTACT HUMAN MAX-DNA COMPLEX: NEW INSIGHTS INTO MECHANISMS OF TRANSCRIPTIONAL CONTROL | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*CP*CP*AP*CP*GP*TP*GP*GP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR MAX) | | Authors: | Brownlie, P, Ceska, T.A, Lamers, M, Romier, C, Theo, H, Suck, D. | | Deposit date: | 1997-09-10 | | Release date: | 1997-10-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control.
Structure, 5, 1997
|
|
