3CDK
 
 | | Crystal structure of the co-expressed succinyl-CoA transferase A and B complex from Bacillus subtilis | | 分子名称: | Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit A, Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit B | | 著者 | Kim, Y, Zhou, M, Stols, L, Eschenfeldt, W, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-02-27 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal structure of the co-expressed succinyl-CoA transferase A and B complex from Bacillus subtilis.
To be Published
|
|
6CP8
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CdiA, CdiI, ... | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2018-03-13 | | 公開日 | 2019-03-13 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
3TO3
 
 | | Crystal Structure of Petrobactin Biosynthesis Protein AsbB from Bacillus anthracis str. Sterne | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Kim, Y, Eschenfeldt, W, Stols, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-09-03 | | 公開日 | 2011-10-05 | | 最終更新日 | 2012-06-06 | | 実験手法 | X-RAY DIFFRACTION (2.382 Å) | | 主引用文献 | Functional and Structural Analysis of the Siderophore Synthetase AsbB through Reconstitution of the Petrobactin Biosynthetic Pathway from Bacillus anthracis.
J.Biol.Chem., 287, 2012
|
|
6WEN
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in the apo form | | 分子名称: | CHLORIDE ION, Non-structural protein 3 | | 著者 | Michalska, K, Stols, L, Jedrzejczak, R, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-02 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
5FFP
 
 | | Crystal structure of CdiI from Burkholderia dolosa AUO158 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Immunity 23 family protein | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2015-12-18 | | 公開日 | 2016-01-20 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of CdiI from Burkholderia dolosa AUO158
To Be Published
|
|
4Q7O
 
 | | The crystal structure of an immunity protein NMB0503 from Neisseria meningitidis MC58 | | 分子名称: | BROMIDE ION, FORMIC ACID, Immunity protein | | 著者 | Tan, K, Stols, L, Eschenfeldt, W, Babnigg, G, Low, D.A, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2014-04-25 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The structure of a contact-dependent growth-inhibition (CDI) immunity protein from Neisseria meningitidis MC58.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
1T8Q
 
 | | Structural genomics, Crystal structure of Glycerophosphoryl diester phosphodiesterase from E. coli | | 分子名称: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, periplasmic, ... | | 著者 | Zhang, R, Kim, Y, Dementieva, I, Duke, N, Stols, L, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-05-13 | | 公開日 | 2004-08-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of Glycerophosphoryl diester phosphodiesterase from E. coli
To be Published
|
|
6CP9
 
 | | Contact-dependent growth inhibition toxin - immunity protein complex from Klebsiella pneumoniae 342 | | 分子名称: | CdiA, CdiI | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2018-03-13 | | 公開日 | 2019-03-13 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
4KT7
 
 | | The crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritolsynthase from Anaerococcus prevotii DSM 20548 | | 分子名称: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, CHLORIDE ION, SODIUM ION | | 著者 | Borek, D, Tan, K, Stols, L, Eschenfeidt, W.H, Otwinoski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-05-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | The crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritolsynthase from Anaerococcus prevotii DSM 20548
To be Published
|
|
5T86
 
 | | Crystal structure of CDI complex from E. coli A0 34/86 | | 分子名称: | ACETATE ION, CdiA toxin, CdiI immunity protein | | 著者 | Michalska, K, Stols, L, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-09-06 | | 公開日 | 2017-09-13 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of CDI complex from E. coli A0 34/86
To Be Published
|
|
4H3T
 
 | | Crystal structure of CRISPR-associated protein Cse1 from Acidimicrobium ferrooxidans | | 分子名称: | CRISPR-associated protein, Cse1 family, GLYCEROL | | 著者 | Michalska, K, Stols, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-09-14 | | 公開日 | 2012-09-26 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structure of CRISPR-associated protein Cse1 from Acidimicrobium ferrooxidans
To be Published
|
|
8EY4
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from E. coli O32:H37 | | 分子名称: | Cys_rich_CPCC domain-containing protein, FE (III) ION, PT-VENN domain-containing protein | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-10-26 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Contact-dependent growth inhibition toxin-immunity protein complex from E. coli O32:H37
To Be Published
|
|
8EY3
 
 | | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37 | | 分子名称: | Cys_rich_CPCC domain-containing protein, FE (III) ION, SODIUM ION | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-10-26 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37
To Be Published
|
|
7M5F
 
 | | Contact-dependent inhibition system from Serratia marcescens BWH57 | | 分子名称: | CdiI, MALONATE ION, Toxin CdiA | | 著者 | Michalska, K, Nutt, W, Stols, L, Jedrzejczak, R, Hayes, C.S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-03-23 | | 公開日 | 2021-05-12 | | 最終更新日 | 2021-08-18 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Contact-dependent inhibition system from Serratia marcescens
To Be Published
|
|
6VEK
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006, full-length | | 分子名称: | contact-dependent immunity protein CdiI, contact-dependent toxin CdiA | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-01-02 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006, full-length
To Be Published
|
|
6W9C
 
 | | The crystal structure of papain-like protease of SARS CoV-2 | | 分子名称: | CHLORIDE ION, Non-structural protein 3, ZINC ION | | 著者 | Osipiuk, J, Jedrzejczak, R, Tesar, C, Endres, M, Stols, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-22 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of papain-like protease of SARS CoV-2
to be published
|
|
6WQF
 
 | |
6WCF
 
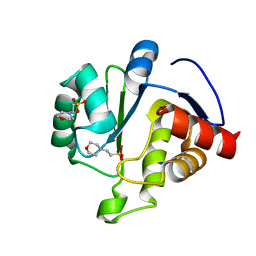 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural protein 3 | | 著者 | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-30 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.065 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
5I4Q
 
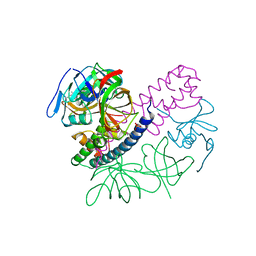 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (domains 2 and 3) | | 分子名称: | CHLORIDE ION, Contact-dependent inhibitor A, Contact-dependent inhibitor I, ... | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2016-02-12 | | 公開日 | 2017-06-28 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
5I4R
 
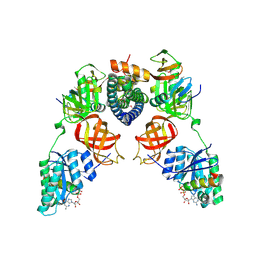 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (trypsin-modified) | | 分子名称: | Contact-dependent inhibitor A, Contact-dependent inhibitor I, Elongation factor Tu, ... | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2016-02-12 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
5HKQ
 
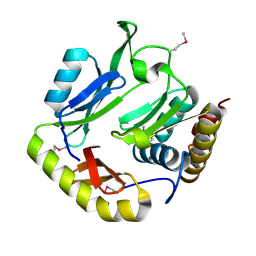 | | Crystal structure of CDI complex from Escherichia coli STEC_O31 | | 分子名称: | CdiI immunity protein, Contact-dependent inhibitor A | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | 登録日 | 2016-01-14 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-03-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional plasticity of antibacterial EndoU toxins.
Mol.Microbiol., 109, 2018
|
|
7OC9
 
 | |
3F0C
 
 | | Crystal structure of transcriptional regulator from Cytophaga hutchinsonii ATCC 33406 | | 分子名称: | SULFATE ION, Transcriptional regulator | | 著者 | Nocek, B, Maltseva, N, Tan, K, Abdullah, J, Eschenfeldt, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-10-24 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Crystal structure of transcriptional regulator from Cytophaga hutchinsonii ATCC 33406
To be Published
|
|
7M6B
 
 | | The Crystal Structure of Mcbe1 | | 分子名称: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | 著者 | Alahuhta, P.M, Lunin, V.V. | | 登録日 | 2021-03-25 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | 著者 | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-18 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
