4BTQ
 
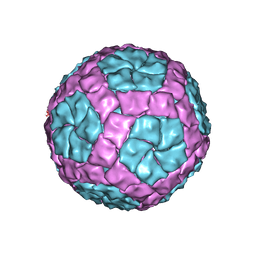 | | Coordinates of the bacteriophage phi6 capsid subunits fitted into the cryoEM map EMD-1206 | | Descriptor: | MAJOR INNER PROTEIN P1 | | Authors: | Nemecek, D, Boura, E, Wu, W, Cheng, N, Plevka, P, Qiao, J, Mindich, L, Heymann, J.B, Hurley, J.H, Steven, A.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Subunit Folds and Maturation Pathway of a Dsrna Virus Capsid.
Structure, 21, 2013
|
|
4BT1
 
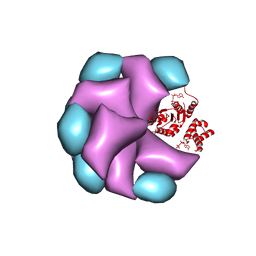 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BT0
 
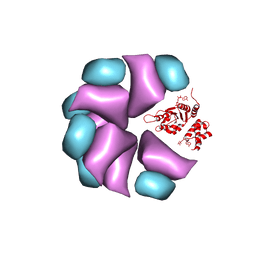 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BTG
 
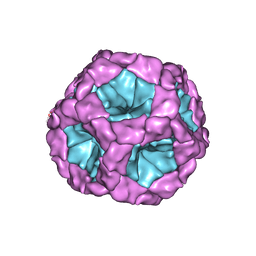 | | Coordinates of the bacteriophage phi6 capsid subunits (P1A and P1B) fitted into the cryoEM reconstruction of the procapsid at 4.4 A resolution | | Descriptor: | MAJOR INNER PROTEIN P1 | | Authors: | Nemecek, D, Boura, E, Wu, W, Cheng, N, Plevka, P, Qiao, J, Mindich, L, Heymann, J.B, Hurley, J.H, Steven, A.C. | | Deposit date: | 2013-06-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Subunit Folds and Maturation Pathway of a Dsrna Virus Capsid.
Structure, 21, 2013
|
|
6UI7
 
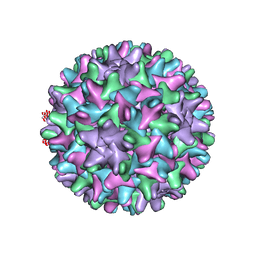 | | HBV T=4 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
6UI6
 
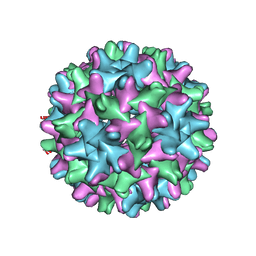 | | HBV T=3 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
6QJT
 
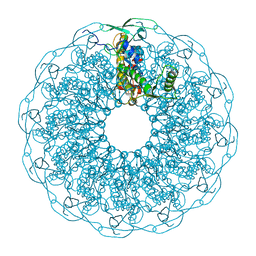 | |
4K7H
 
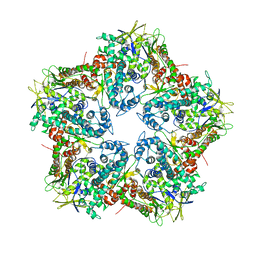 | | Major capsid protein P1 of the Pseudomonas phage phi6 | | Descriptor: | Major inner protein P1 | | Authors: | Boura, E, Nemecek, D, Plevka, P, Steven, C.A, Hurley, J.H. | | Deposit date: | 2013-04-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5964 Å) | | Cite: | Subunit Folds and Maturation Pathway of a dsRNA Virus Capsid.
Structure, 21, 2013
|
|
4D1Q
 
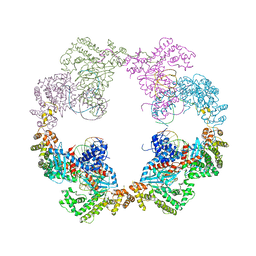 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
6CF2
 
 | | Crystal structure of HIV-1 Rev (residues 1-93)-RNA aptamer complex | | Descriptor: | Anti-Rev Antibody, heavy chain, light chain, ... | | Authors: | Eren, E, Dearborn, A.D, Wingfield, P.T. | | Deposit date: | 2018-02-13 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an RNA Aptamer that Can Inhibit HIV-1 by Blocking Rev-Cognate RNA (RRE) Binding and Rev-Rev Association.
Structure, 26, 2018
|
|
1C5E
 
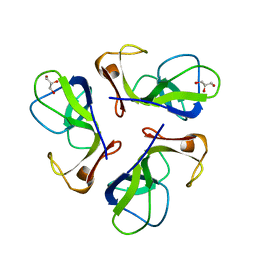 | | BACTERIOPHAGE LAMBDA HEAD PROTEIN D | | Descriptor: | GLYCEROL, HEAD DECORATION PROTEIN | | Authors: | Yang, F, Forrer, P, Dauter, Z, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1999-11-18 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD.
Nat.Struct.Biol., 7, 2000
|
|
6AXW
 
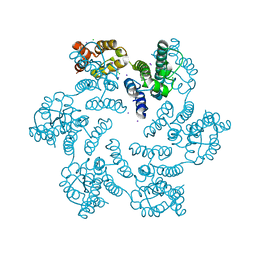 | |
6AX5
 
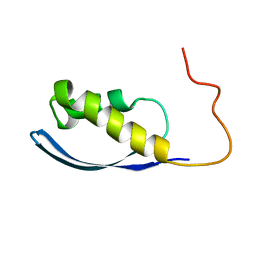 | | RPT1 region of INI1/SNF5/SMARCB1_HUMAN - SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Girvin, M.E, Cahill, S.M, Harris, R, Cowburn, D, Spira, M, Wu, X, Prakash, R, Bernowitz, M, Almo, S.C, Kalpana, G.V. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | INI1/SMARCB1 Rpt1 domain mimics TAR RNA in binding to integrase to facilitate HIV-1 replication.
Nat Commun, 12, 2021
|
|
6AXR
 
 | |
3V8U
 
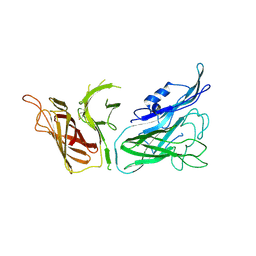 | |
3V89
 
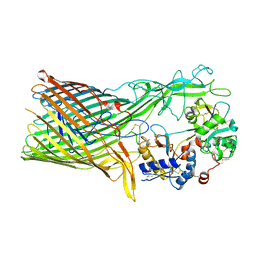 | | The crystal structure of transferrin binding protein A (TbpA) from Neisseria meningitidis serogroup B in complex with the C-lobe of human transferrin | | Descriptor: | Serotransferrin, Transferrin-binding protein A | | Authors: | Noinaj, N, Oke, M, Easley, N, Zak, O, Aisen, P, Buchanan, S.K. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-15 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for iron piracy by pathogenic Neisseria.
Nature, 483, 2012
|
|
3V83
 
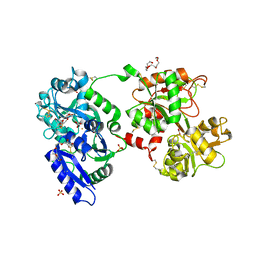 | | The 2.1 angstrom crystal structure of diferric human transferrin | | Descriptor: | BICARBONATE ION, FE (III) ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Noinaj, N, Steere, A, Mason, A.B, Buchanan, S.K. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-15 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural basis for iron piracy by pathogenic Neisseria.
Nature, 483, 2012
|
|
3V8X
 
 | | The crystal structure of transferrin binding protein A (TbpA) from Neisserial meningitidis serogroup B in complex with full length human transferrin | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Noinaj, N, Easley, N, Buchanan, S.K. | | Deposit date: | 2011-12-23 | | Release date: | 2012-02-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for iron piracy by pathogenic Neisseria.
Nature, 483, 2012
|
|
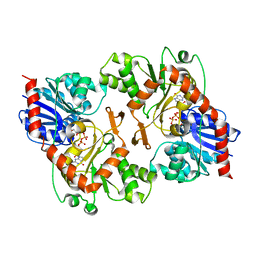
3OY7
 
 | |
3OY2
 
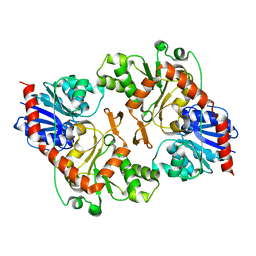 | |
4EPA
 
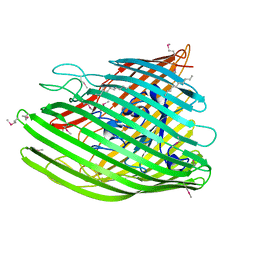 | |
4EPI
 
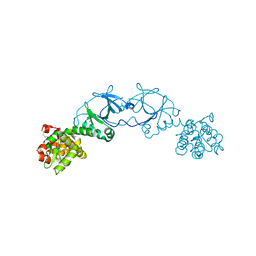 | | The crystal structure of pesticin-T4 lysozyme hybrid stabilized by engineered disulfide bonds | | Descriptor: | Pesticin, Lysozyme Chimera, SODIUM ION, ... | | Authors: | Seddiki, N, Fairman, J.W, Noinaj, N, Lukacik, P, Barnard, T, Buchanan, S.K. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural engineering of a phage lysin that targets Gram-negative pathogens.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EPF
 
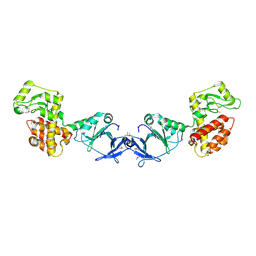 | |
4EXM
 
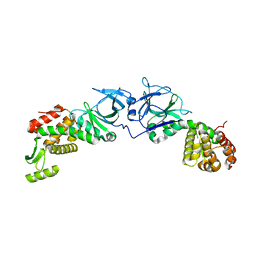 | | The crystal structure of an engineered phage lysin containing the binding domain of pesticin and the killing domain of T4-lysozyme | | Descriptor: | Pesticin, Lysozyme Chimera | | Authors: | Seddiki, N, Noinaj, N, Fairman, J.W, Lukacik, P, Barnard, T.J, Buchanan, S.K. | | Deposit date: | 2012-04-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural engineering of a phage lysin that targets Gram-negative pathogens.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G1G
 
 | |
