3KL3
 
 | | Crystal structure of Ligand bound XynC | | Descriptor: | D-HISTIDINE, Glucuronoxylanase xynC, TETRAETHYLENE GLYCOL, ... | | Authors: | St John, F.J, Hurlbert, J.C, Pozharski, E. | | Deposit date: | 2009-11-06 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase.
J.Mol.Biol., 407, 2011
|
|
3KL0
 
 | | Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis | | Descriptor: | D(-)-TARTARIC ACID, Glucuronoxylanase xynC, HISTIDINE, ... | | Authors: | St John, F.J, Hurlbert, J.C, Pozharski, E. | | Deposit date: | 2009-11-06 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase.
J.Mol.Biol., 407, 2011
|
|
3KL5
 
 | |
3GTN
 
 | |
3RDK
 
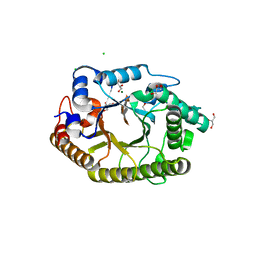 | | Protein crystal structure of xylanase A1 of Paenibacillus sp. JDR-2 | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose, CHLORIDE ION, Endo-1,4-beta-xylanase, ... | | Authors: | Pozharski, E, St John, F.J. | | Deposit date: | 2011-04-01 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel structural features of xylanase A1 from Paenibacillus sp. JDR-2.
J.Struct.Biol., 180, 2012
|
|
3RO8
 
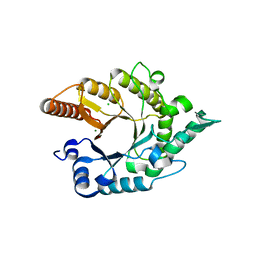 | |
4FMV
 
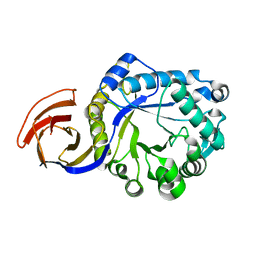 | | Crystal Structure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 | | Descriptor: | Glucuronoarabinoxylan endo-1,4-beta-xylanase | | Authors: | Bales, E.B, Smith, J.K, St John, F.J, Hurlbert, J.C. | | Deposit date: | 2012-06-18 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel member of glycoside hydrolase family 30 subfamily 8 with altered substrate specificity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4E4P
 
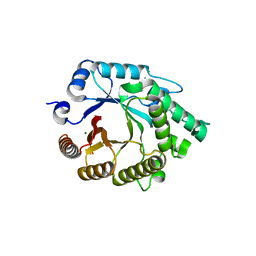 | |
7N6H
 
 | |
6CSJ
 
 | |
5CXP
 
 | | X-ray crystallographic protein structure of the glycoside hydrolase family 30 subfamily 8 xylanase, Xyn30A, from Clostridium acetobutylicum | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Possible xylan degradation enzyme (Glycosyl hydrolase family 30-like domain and Ricin B-like domain), ... | | Authors: | St John, F.J, Pozharski, E, Hurlbert, J.C. | | Deposit date: | 2015-07-29 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a GH30 xylanase
To Be Published
|
|
7N6O
 
 | |
