3GS1
 
 | | An all-RNA Hairpin Ribozyme with mutation A38N1dA | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
3GS5
 
 | | An all-RNA hairpin ribozyme A38N1dA variant with a product mimic substrate strand | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (25-MER), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
3GS8
 
 | | An all-RNA hairpin ribozyme A38N1dA38 variant with a transition-state mimic substrate strand | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
3GCA
 
 | |
3I2R
 
 | | Crystal structure of the hairpin ribozyme with a 2',5'-linked substrate with N1-deazaadenosine at position A9 | | Descriptor: | 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
3I2S
 
 | | Crystal structure of the hairpin ribozyme with a 2'OMe substrate and N1-deazaadenosine at position A10 | | Descriptor: | 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
3I2Q
 
 | | Crystal structure of the hairpin ribozyme with 2'OMe substrate strand and N1-deazaadenosine at position A9 | | Descriptor: | 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
3I2U
 
 | | Crystal structure of the haiprin ribozyme with a 2',5'-linked substrate and N1-deazaadenosine at position A10 | | Descriptor: | 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
6N55
 
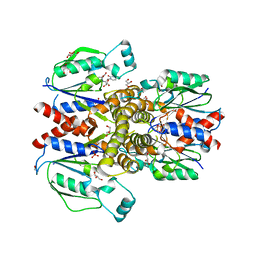 | | Crystal structure of human uridine-cytidine kinase 2 complexed with 2'-azidouridine | | Descriptor: | 2'-azido-2'-deoxyuridine, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Cuthbert, B.J, Nainar, S, Spitale, R.C, Goulding, C.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.085 Å) | | Cite: | An optimized chemical-genetic method for cell-specific metabolic labeling of RNA.
Nat.Methods, 17, 2020
|
|
6N54
 
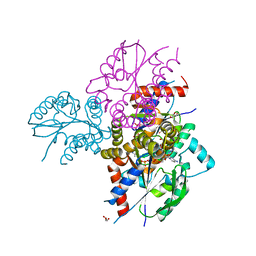 | | Crystal structure of human uridine-cytidine kinase 2 complexed with 2'-azidocytidine monophosphate | | Descriptor: | 2'-azido-2'-deoxycytidine 5'-(dihydrogen phosphate), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuthbert, B.J, Nainar, S, Spitale, R.C, Goulding, C.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | An optimized chemical-genetic method for cell-specific metabolic labeling of RNA.
Nat.Methods, 17, 2020
|
|
6N53
 
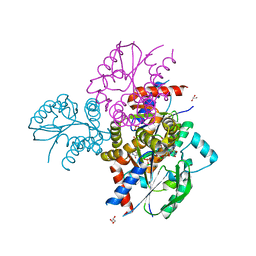 | | Crystal structure of human uridine-cytidine kinase 2 complexed with 2'-azidouridine monophosphate | | Descriptor: | 2'-deoxy-2'-triaza-1,2-dien-2-ium-1-yl-uridine-5'-monophosphate, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuthbert, B.J, Nainar, S, Spitale, R.C, Goulding, C.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An optimized chemical-genetic method for cell-specific metabolic labeling of RNA.
Nat.Methods, 17, 2020
|
|
3CQS
 
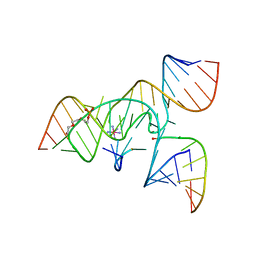 | | A 3'-OH, 2',5'-phosphodiester substitution in the hairpin ribozyme active site reveals similarities with protein ribonucleases | | Descriptor: | 13-mer substrate strand with 3'-OH, 2',5'-phosphodiester covalently linking 5th and 6th nucleotides, 19-mer ribozyme strand, ... | | Authors: | Torelli, A.T, Spitale, R.C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Shared traits on the reaction coordinates of ribonuclease and an RNA enzyme
Biochem.Biophys.Res.Commun., 371, 2008
|
|
6PWZ
 
 | |
4RZD
 
 | | Crystal Structure of a PreQ1 Riboswitch | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, PreQ1-III Riboswitch (Class 3) | | Authors: | Wedekind, J.E, Liberman, J.A, Salim, M. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2NPY
 
 | | Crystal Structure of a junctioned hairpin ribozyme incorporating 9atom linker and 2'-deoxy 2'-amino U at A-1 | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2006-10-30 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A posteriori design of crystal contacts to improve the X-ray diffraction properties of a small RNA enzyme.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2NPZ
 
 | | Crystal structure of junctioned hairpin ribozyme incorporating synthetic propyl linker | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2006-10-30 | | Release date: | 2007-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A posteriori design of crystal contacts to improve the X-ray diffraction properties of a small RNA enzyme.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3Q51
 
 | |
3Q50
 
 | |
