6A1U
 
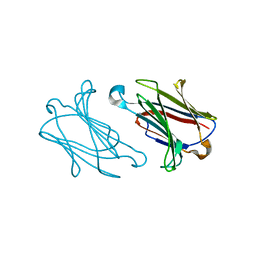 | | Charcot-Leyden crystal protein/Galectin-10 variant E33D | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
6A64
 
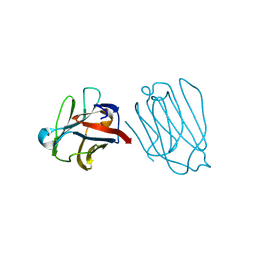 | | Placental protein 13/galectin-13 variant R53HR55NH57RD33G with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A1Y
 
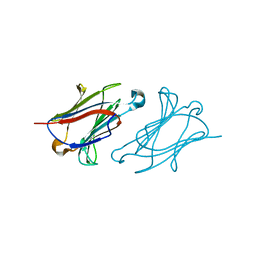 | | Charcot-Leyden crystal protein/Galectin-10 variant Y35A | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
6A65
 
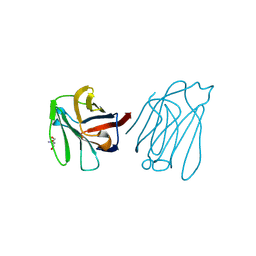 | | Placental protein 13/galectin-13 variant R53HR55N with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A1V
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant E33Q | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
6A63
 
 | | Placental protein 13/galectin-13 variant R53HH57R with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A66
 
 | | Placental protein 13/galectin-13 variant R53H with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J.Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A1S
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant E33A | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
6A1X
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant W127A | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
7E0H
 
 | | LHCII-1 in the state transition supercomplex PSI-LHCI-LHCII from the LhcbM1 lacking mutant of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7DZ8
 
 | | State transition supercomplex PSI-LHCI-LHCII from the LhcbM1 lacking mutant of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-23 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7E0J
 
 | | LHCII-1 in the state transition supercomplex PSI-LHCI-LHCII from the double phosphatase mutant pph1;pbcp of Chlamydomonas reinhardti. | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7DZ7
 
 | | State transition supercomplex PSI-LHCI-LHCII from double phosphatase mutant pph1;pbcp of green alga Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-23 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7E0K
 
 | | LHCII-2 in the state transition supercomplex PSI-LHCI-LHCII from the double phosphatase mutant pph1;pbcp of Chlamydomonas reinhardti. | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7E0I
 
 | | LHCII-2 in the state transition supercomplex PSI-LHCI-LHCII from the LhcbM1 lacking mutant of Chlamydomonas reinhardti | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | Deposit date: | 2021-01-28 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7XHA
 
 | |
7XHB
 
 | |
7YDO
 
 | | Crystal structure of Atg44 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Uncharacterized protein C26A3.14c | | Authors: | Maruyama, T, Noda, N.N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-17 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The mitochondrial intermembrane space protein mitofissin drives mitochondrial fission required for mitophagy.
Mol.Cell, 83, 2023
|
|
8AYP
 
 | | NaK C-DI mutant with Rb+ and Ba2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BARIUM ION, Potassium channel protein, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Calcium Permeation in a Glutamate Receptor Ion Channel.
J.Chem.Inf.Model., 63, 2023
|
|
8AYQ
 
 | | NaK C-DI mutant with Rb+ and Ca2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanism of Calcium Permeation in a Glutamate Receptor Ion Channel.
J.Chem.Inf.Model., 63, 2023
|
|
