6IUN
 
 | |
6IUM
 
 | |
8JB1
 
 | |
5IWQ
 
 | |
8HRR
 
 | |
8HRT
 
 | |
8HRO
 
 | |
8HRQ
 
 | |
8HRS
 
 | |
8HRP
 
 | |
5Z6U
 
 | |
5Z6T
 
 | |
8GQM
 
 | |
8GQK
 
 | |
8GQI
 
 | |
8GQJ
 
 | |
8GQF
 
 | |
8GQH
 
 | |
8GQN
 
 | |
8GQL
 
 | |
8GQG
 
 | |
5L2D
 
 | |
3HRT
 
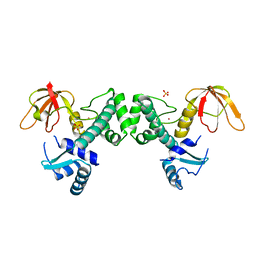 | | Crystal Structure of ScaR with bound Cd2+ | | Descriptor: | CADMIUM ION, Metalloregulator ScaR, SULFATE ION | | Authors: | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
3HRS
 
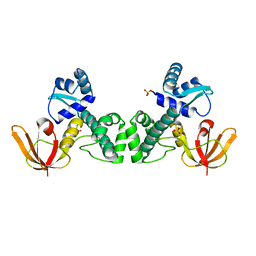 | | Crystal Structure of the Manganese-activated Repressor ScaR: apo form | | Descriptor: | Metalloregulator ScaR, SULFATE ION | | Authors: | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
3HRU
 
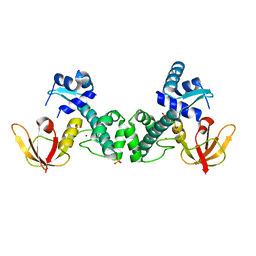 | | Crystal Structure of ScaR with bound Zn2+ | | Descriptor: | Metalloregulator ScaR, SULFATE ION, ZINC ION | | Authors: | Stoll, K.E, Draper, W.E, Kliegman, J.I, Golynskiy, M.V, Brew-Appiah, R.A.T, Brown, H.K, Breyer, W.A, Jakubovics, N.S, Jenkinson, H.F, Brennan, R.B, Cohen, S.M, Glasfeld, A. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization and structure of the manganese-responsive transcriptional regulator ScaR.
Biochemistry, 48, 2009
|
|
