8AUG
 
 | |
8AU8
 
 | |
8AUA
 
 | |
8AUH
 
 | |
8AUL
 
 | |
8AU9
 
 | |
4LGX
 
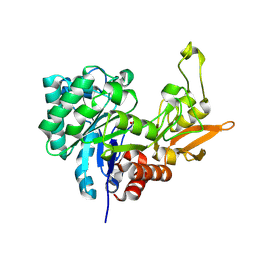 | | Structure of Chitinase D from Serratia proteamaculans revealed an unusually constrained substrate binding site | | Descriptor: | ACETATE ION, GLYCEROL, Glycoside hydrolase family 18 | | Authors: | Madhuprakash, J, Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2013-06-30 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Inverse relationship between chitobiase and transglycosylation activities of chitinase-D from Serratia proteamaculans revealed by mutational and biophysical analyses.
Sci Rep, 5, 2015
|
|
3SVS
 
 | |
3SVO
 
 | |
3SVN
 
 | |
3SVU
 
 | |
3SVR
 
 | |
4MLE
 
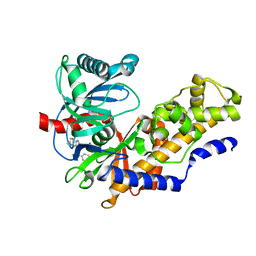 | | Human Glucokinase in Complex with Novel Amino Thiazole Activator | | Descriptor: | 3-(benzyloxy)-N-(4-methyl-1,3-thiazol-2-yl)pyridin-2-amine, Glucokinase, alpha-D-glucopyranose | | Authors: | Voegtli, W.C. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a New Class of Glucokinase Activators through Structure-Based Design.
J.Med.Chem., 56, 2013
|
|
4MLH
 
 | |
7LNK
 
 | |
7F5Z
 
 | | Crystal structure of the single-stranded dna-binding protein from Mycobacterium tuberculosis- Form III | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Srikalaivani, R, Paul, A, Sriram, R, Narayanan, S, Gopal, B, Vijayan, M. | | Deposit date: | 2021-06-23 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural variability of Mycobacterium tuberculosis SSB and susceptibility to inhibition.
Curr.Sci., 122, 2022
|
|
7F5Y
 
 | | Crystal structure of the single-stranded dna-binding protein from Mycobacterium tuberculosis- Form III | | Descriptor: | FORMIC ACID, Single-stranded DNA-binding protein | | Authors: | Srikalaivani, R, Paul, A, Sriram, R, Narayanan, S, Gopal, B, Vijayan, M. | | Deposit date: | 2021-06-23 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural variability of Mycobacterium tuberculosis SSB and susceptibility to inhibition.
Curr.Sci., 122, 2022
|
|
7JHI
 
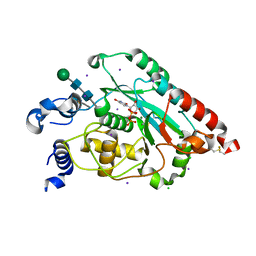 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 iodide-derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7JHM
 
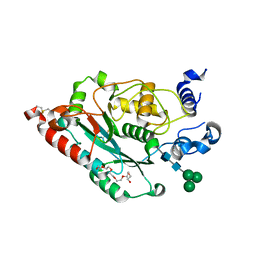 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEXAETHYLENE GLYCOL, N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase 2, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7JHO
 
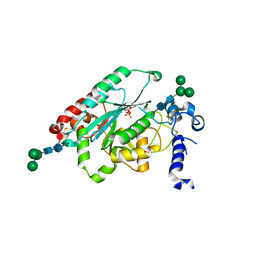 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7JHN
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP and trisaccharide GlcNAc-beta1-3Gal-beta1-4GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7JHK
 
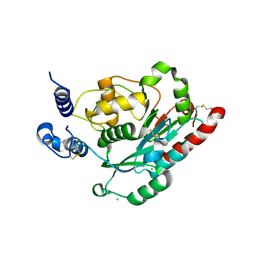 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 in unliganded form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3436 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
7JHL
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP-N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
