2MS3
 
 | |
6N3S
 
 | | Crystal structure of apo-cruzain | | 分子名称: | 1,2-ETHANEDIOL, Cruzipain, PHOSPHATE ION | | 著者 | Silva, E.B, Dall, E, Rodrigues, F.T.G, Ferreira, R.S, Brandstetter, H. | | 登録日 | 2018-11-16 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.193 Å) | | 主引用文献 | Cruzain structures: apocruzain and cruzain bound to S-methyl thiomethanesulfonate and implications for drug design.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6O2X
 
 | | Structure of cruzain bound to MMTS inhibitor | | 分子名称: | 1,2-ETHANEDIOL, Cruzipain, PHOSPHATE ION | | 著者 | Silva, E.B, Dall, E, Ferreira, R.S, Brandstetter, H. | | 登録日 | 2019-02-25 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.193 Å) | | 主引用文献 | Cruzain structures: apocruzain and cruzain bound to S-methyl thiomethanesulfonate and implications for drug design.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7JUJ
 
 | | Cruzain bound to Gallinamide inhibitor | | 分子名称: | Cruzipain, POTASSIUM ION, gallinamide A, ... | | 著者 | Silva, E.B, Sharma, V, Alvarez, L.H, Gerwick, W.H, McKerrow, J.H, Podust, L.M. | | 登録日 | 2020-08-19 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Intramolecular Interactions Enhance the Potency of Gallinamide A Analogues against Trypanosoma cruzi .
J.Med.Chem., 65, 2022
|
|
3IGM
 
 | | A 2.2A crystal structure of the AP2 domain of PF14_0633 from P. falciparum, bound as a domain-swapped dimer to its cognate DNA | | 分子名称: | 5'-D(*TP*GP*CP*AP*TP*GP*CP*A)-3', PF14_0633 protein | | 著者 | Lindner, S.E, De Silva, E, Keck, J.L, Llinas, M. | | 登録日 | 2009-07-28 | | 公開日 | 2009-11-10 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Determinants of DNA Binding by a P. falciparum ApiAP2 Transcriptional Regulator.
J.Mol.Biol., 395, 2010
|
|
5LMC
 
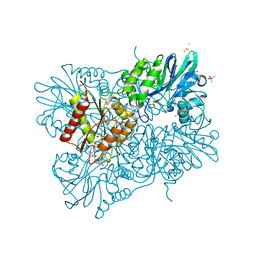 | | Oxidized flavodiiron core of Escherichia coli flavorubredoxin, including the Fe-4SG atoms from its rubredoxin domain | | 分子名称: | ACETIC ACID, Anaerobic nitric oxide reductase flavorubredoxin, CACODYLATE ION, ... | | 著者 | Romao, C.V, Borges, P.T, Vicente, J.B, Carrondo, M.A, Teixeira, M, Frazao, C. | | 登録日 | 2016-07-29 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Escherichia coli Flavodiiron Nitric Oxide Reductase.
J.Mol.Biol., 428, 2016
|
|
5LLD
 
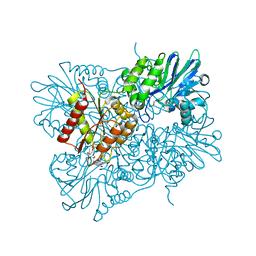 | | Flavodiiron core of Escherichia coli flavorubredoxin in the reduced form. | | 分子名称: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Romao, C.V, Borges, P.T, Vicente, J.B, Carrondo, M.A, Teixeira, M, Frazao, C. | | 登録日 | 2016-07-27 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.651 Å) | | 主引用文献 | Structure of Escherichia coli Flavodiiron Nitric Oxide Reductase.
J.Mol.Biol., 428, 2016
|
|
7X6K
 
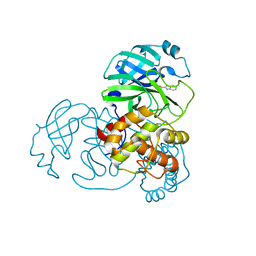 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3w | | 分子名称: | 1H-indole-2-carbaldehyde, 3C-like proteinase | | 著者 | Su, H, Nie, T, Xie, H, Li, Z.W, Li, M.J, Xu, Y. | | 登録日 | 2022-03-07 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination.
J.Med.Chem., 65, 2022
|
|
7X6J
 
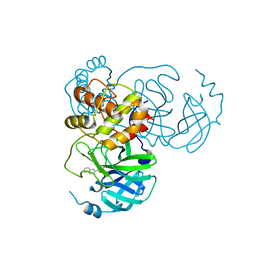 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3af | | 分子名称: | 3C-like proteinase, quinoline-2-carboxylic acid | | 著者 | Su, H, Nie, T, Xie, H, Li, Z.W, Li, M.J, Xu, Y. | | 登録日 | 2022-03-07 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination.
J.Med.Chem., 65, 2022
|
|
6URA
 
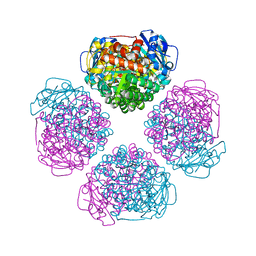 | | Crystal structure of RUBISCO from Promineofilum breve | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain | | 著者 | Pereira, J.H, Banda, D.M, Liu, A.K, Shih, P.M, Adams, P.D. | | 登録日 | 2019-10-23 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Novel bacterial clade reveals origin of form I Rubisco.
Nat.Plants, 6, 2020
|
|
4D02
 
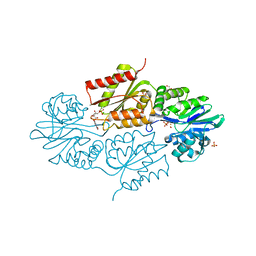 | | The crystallographic structure of Flavorubredoxin from Escherichia coli | | 分子名称: | ANAEROBIC NITRIC OXIDE REDUCTASE FLAVORUBREDOXIN, CHLORIDE ION, FE (III) ION, ... | | 著者 | Romao, C.V, Vicente, J.B, Bandeiras, T, Carrondo, M.A, Teixeira, M, Frazao, C. | | 登録日 | 2014-04-23 | | 公開日 | 2014-09-03 | | 最終更新日 | 2019-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.755 Å) | | 主引用文献 | Structure of Escherichia Coli Flavodiiron Nitric Oxide Reductase.
J.Mol.Biol., 428, 2016
|
|
7S19
 
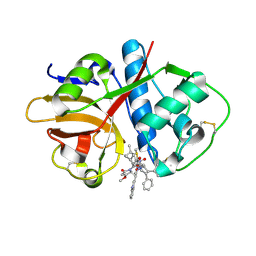 | |
7S18
 
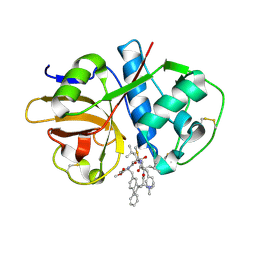 | | Crystal structure of cruzain with gallinamide analog from 2-biaryl series | | 分子名称: | Cruzipain, N,N-dimethyl-L-valyl-L-leucyl-N-[(3S)-6-{(2S)-2-[([1,1'-biphenyl]-4-yl)methyl]-3-methoxy-5-oxo-2,5-dihydro-1H-pyrrol-1-yl}-6-oxo-1-phenylhexan-3-yl]-L-leucinamide | | 著者 | Sharma, V, Podust, L.M. | | 登録日 | 2021-09-01 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Intramolecular Interactions Enhance the Potency of Gallinamide A Analogues against Trypanosoma cruzi .
J.Med.Chem., 65, 2022
|
|
8E1Y
 
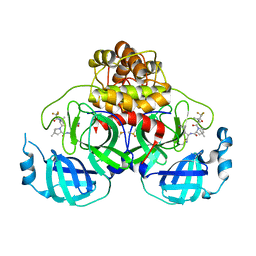 | | Crystal Structure of SARS-CoV-2 Main protease A193S mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ9
 
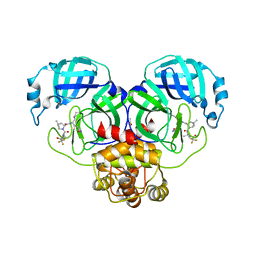 | | Crystal Structure of SARS-CoV-2 Main protease G143S mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.664 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZA
 
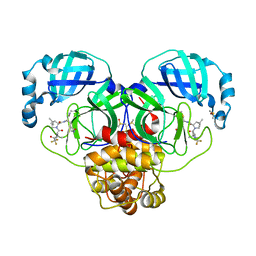 | | Crystal Structure of SARS-CoV-2 Main protease A193T mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.961 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ6
 
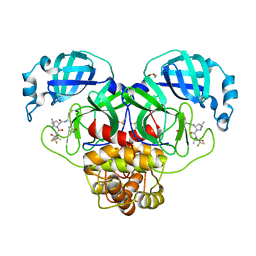 | | Crystal Structure of SARS-CoV-2 Main protease mutant Q189K in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.366 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ0
 
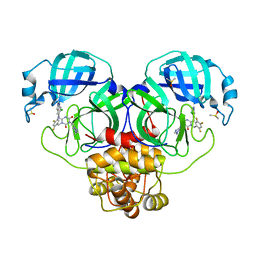 | | Crystal Structure of SARS-CoV-2 Main protease in complex with Ensitrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ1
 
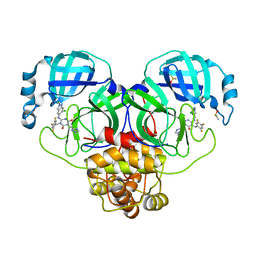 | | Crystal Structure of SARS-CoV-2 Main protease mutant M49I in complex with Ensitrelvir | | 分子名称: | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ2
 
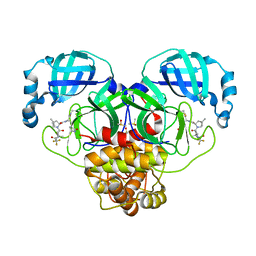 | | Crystal Structure of SARS-CoV-2 Main protease in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.129 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8E26
 
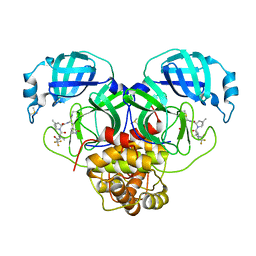 | | Crystal Structure of SARS-CoV-2 Main Protease N142S mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Godoy, A.S, Oliva, G. | | 登録日 | 2022-08-14 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.845 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8E25
 
 | | Crystal Structure of SARS-CoV-2 Main Protease M49I mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab | | 著者 | Noske, G.D, Godoy, A.S, Oliva, G. | | 登録日 | 2022-08-14 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.868 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
