5YMA
 
 | | Crystal structure of ribosome assembly factor Efg1 | | Descriptor: | Putative rRNA processing protein | | Authors: | Shu, S, Ye, K. | | Deposit date: | 2017-10-21 | | Release date: | 2018-01-17 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Structural and functional analysis of ribosome assembly factor Efg1.
Nucleic Acids Res., 46, 2018
|
|
5UJ0
 
 | |
7T6D
 
 | | CryoEM structure of the YejM/LapB complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Inner membrane protein YejM, ... | | Authors: | Mi, W, Shu, S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli.
Nat Commun, 13, 2022
|
|
8V24
 
 | | LapB cytoplasmic domain in complex with LpxC | | Descriptor: | ACETATE ION, Lipopolysaccharide assembly protein B, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Mi, W, Shu, S. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dual function of LapB (YciM) in regulating Escherichia coli lipopolysaccharide synthesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6DTQ
 
 | | Maltose bound T. maritima MalE3 | | Descriptor: | MAGNESIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE3 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTT
 
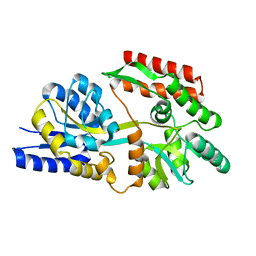 | | Apo T. maritima MalE2 | | Descriptor: | maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6VDD
 
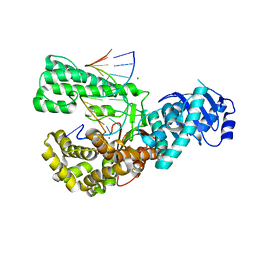 | | POL domain of Pol1 from M. smegmatis complex with DNA primer-template and dNTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A*(DCT))-3'), DNA (5'-D(P*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
6VDE
 
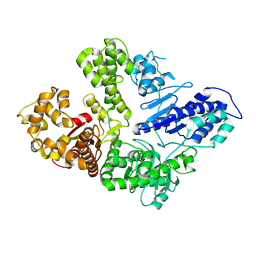 | | Full-length M. smegmatis Pol1 | | Descriptor: | DNA polymerase I, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.713 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
6VDC
 
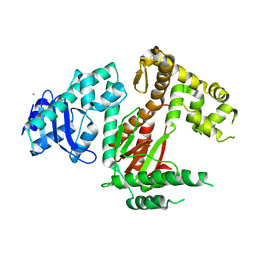 | | POL domain of Pol1 from M. smegmatis | | Descriptor: | DNA polymerase I, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Ghosh, S. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Mycobacterial DNA polymerase I: activities and crystal structures of the POL domain as apoenzyme and in complex with a DNA primer-template and of the full-length FEN/EXO-POL enzyme.
Nucleic Acids Res., 48, 2020
|
|
6NHZ
 
 | | mycobacterial DNA ligase D complexed with ATP and Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent DNA ligase, MAGNESIUM ION | | Authors: | Shuman, S, Unciuleac, M, Goldgur, Y. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of ATP-bound DNA ligase D in a closed domain conformation reveal a network of amino acid and metal contacts to the ATP phosphates.
J. Biol. Chem., 294, 2019
|
|
8DHD
 
 | | Neutron crystal structure of maltotetraose bound tmMBP | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S, Myles, D.A. | | Deposit date: | 2022-06-27 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Mapping periplasmic binding protein oligosaccharide recognition with neutron crystallography.
Sci Rep, 12, 2022
|
|
6DTU
 
 | | Maltotetraose bound T. maritima MalE1 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE1 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6E3A
 
 | | tRNA 2'-phosphotransferase | | Descriptor: | COENZYME A, Probable RNA 2'-phosphotransferase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of tRNA splicing enzyme Tpt1 illuminates the mechanism of RNA 2'-PO4recognition and ADP-ribosylation.
Nat Commun, 10, 2019
|
|
6DTR
 
 | | Apo T. maritima MalE3 | | Descriptor: | SULFATE ION, maltose-binding protein MalE3 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTS
 
 | | Maltotetraose bound T. maritima MalE2 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6EDE
 
 | | tRNA 2'-phosphotransferase | | Descriptor: | Probable RNA 2'-phosphotransferase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structure of tRNA splicing enzyme Tpt1 illuminates the mechanism of RNA 2'-PO4recognition and ADP-ribosylation.
Nat Commun, 10, 2019
|
|
6TZX
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | INOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6U03
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6U00
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6TZO
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6TZM
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-12 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.714 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6C34
 
 | | Mycobacterium smegmatis DNA flap endonuclease mutant D125N | | Descriptor: | 5'-3' exonuclease, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Carl, A, Uson, M.L. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and mutational analysis of Mycobacterium smegmatis FenA highlight active site amino acids and three metal ions essential for flap endonuclease and 5' exonuclease activities.
Nucleic Acids Res., 46, 2018
|
|
6C36
 
 | | Mycobacterium smegmatis flap endonuclease mutant D208N | | Descriptor: | 5'-3' exonuclease, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Shuman, S, Goldgur, Y, Carl, A, Uson, M.L. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mutational analysis of Mycobacterium smegmatis FenA highlight active site amino acids and three metal ions essential for flap endonuclease and 5' exonuclease activities.
Nucleic Acids Res., 46, 2018
|
|
6U05
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
6C35
 
 | | Mycobacterium smegmatis flap endonuclease mutant D148N | | Descriptor: | 5'-3' exonuclease, MANGANESE (II) ION | | Authors: | Shuman, S, Goldgur, Y, Carl, A, Uson, M.L. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal structure and mutational analysis of Mycobacterium smegmatis FenA highlight active site amino acids and three metal ions essential for flap endonuclease and 5' exonuclease activities.
Nucleic Acids Res., 46, 2018
|
|
