3VQT
 
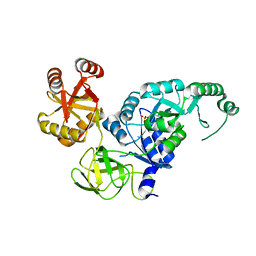 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
3VR1
 
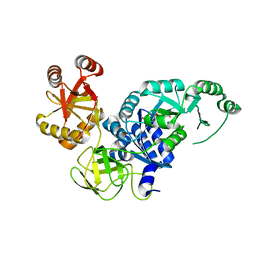 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-04-03 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
3WYO
 
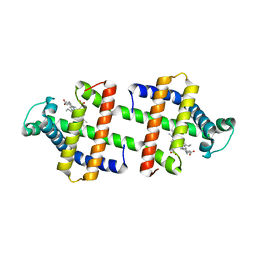 | | Heterodimeric myoglobin formed by domain swapping | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y.W, Nagao, S, Zhang, M, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of heterodimeric protein using domain swapping for myoglobin.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3VM9
 
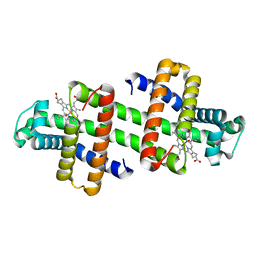 | | Dimeric horse myoglobin | | Descriptor: | Myoglobin, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Osuka, H, Yamada, T, Uni, T, Shomura, Y, Imai, K, Higuchi, Y, Hirota, S. | | Deposit date: | 2011-12-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural and oxygen binding properties of dimeric horse myoglobin
Dalton Trans, 41, 2012
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVB
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | Descriptor: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVC
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XEC
 
 | | Heterodimer constructed from PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
5XED
 
 | | Heterodimer constructed from M61A PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
3ABQ
 
 | |
3ABR
 
 | |
3ABO
 
 | |
3ABS
 
 | |
