5YRH
 
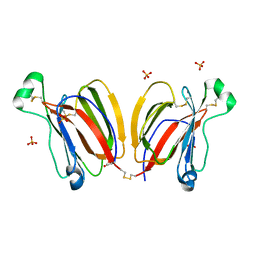 | | Crystal structure of PPL3B | | Descriptor: | PPL3-a, PPL3-b, SULFATE ION | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YRM
 
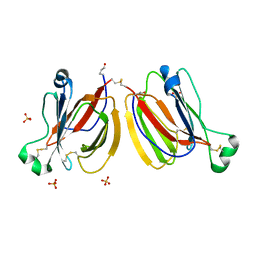 | | PPL3C-isomaltose complex | | Descriptor: | PPL3-b, SULFATE ION, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YT3
 
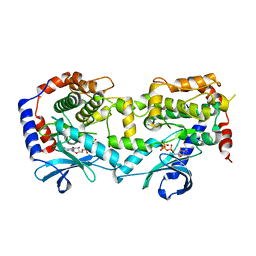 | | Structure of the Human Mitogen-Activated Protein Kinase Kinase 1 S218D and S222D mutant | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase kinase 1, isoform CRA_d, ... | | Authors: | Nakae, S, Doko, K, Tada, T, Shirai, T. | | Deposit date: | 2017-11-16 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Human Mitogen-Activated Protein Kinase Kinase 1 S218D and S222D mutant
To Be Published
|
|
5YRF
 
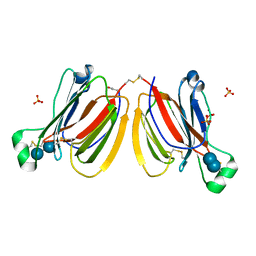 | | PPL3A-trehalose complex | | Descriptor: | PPL3-A, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YRI
 
 | | PPL3B-trehalose complex | | Descriptor: | PPL3-a, PPL3-b, SULFATE ION, ... | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YRJ
 
 | | PPL3B-isomaltose complex | | Descriptor: | PPL3-a, PPL3-b, SULFATE ION, ... | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YRL
 
 | | PPL3C-trehalose complex | | Descriptor: | PPL3-b, SULFATE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YRE
 
 | | Crystal structure of PPL3A | | Descriptor: | GLYCEROL, PPL3-A, SULFATE ION | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YRG
 
 | | PPL3A-isomaltose complex | | Descriptor: | PPL3-A, SULFATE ION, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5GHS
 
 | | DNA replication protein | | Descriptor: | Putative uncharacterized protein, SULFATE ION, SsDNA-specific exonuclease | | Authors: | Oyama, T. | | Deposit date: | 2016-06-20 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication.
Nucleic Acids Res., 44, 2016
|
|
5GHR
 
 | | DNA replication protein | | Descriptor: | Putative uncharacterized protein, SULFATE ION, SsDNA-specific exonuclease | | Authors: | Oyama, T. | | Deposit date: | 2016-06-20 | | Release date: | 2016-10-12 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication.
Nucleic Acids Res., 44, 2016
|
|
5GHT
 
 | | DNA replication protein | | Descriptor: | SsDNA-specific exonuclease | | Authors: | Oyama, T. | | Deposit date: | 2016-06-20 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication.
Nucleic Acids Res., 44, 2016
|
|
2D7H
 
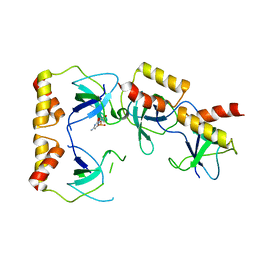 | | Crystal structure of the ccc complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*CP*CP*C)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7G
 
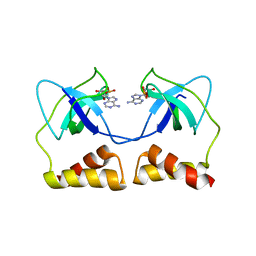 | | Crystal structure of the aa complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*AP*A)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7E
 
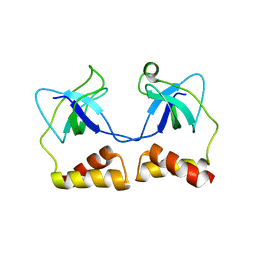 | | Crystal structure of N-terminal domain of PriA from E.coli | | Descriptor: | Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-18 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2ZJ5
 
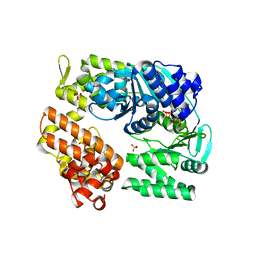 | | Archaeal DNA helicase Hjm complexed with ADP in form 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative ski2-type helicase, SULFATE ION | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
2ZJA
 
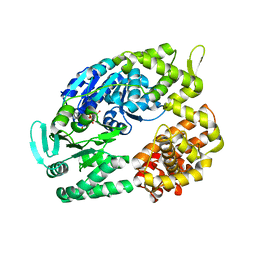 | | Archaeal DNA helicase Hjm complexed with AMPPCP in form 2 | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Putative ski2-type helicase | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
2ZJ2
 
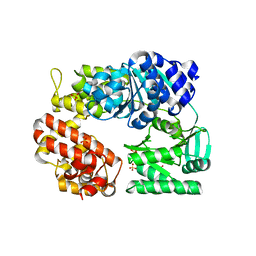 | | Archaeal DNA helicase Hjm apo state in form 1 | | Descriptor: | Putative ski2-type helicase, SULFATE ION | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
2ZJ8
 
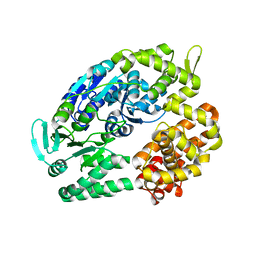 | | Archaeal DNA helicase Hjm apo state in form 2 | | Descriptor: | Putative ski2-type helicase | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
2DWN
 
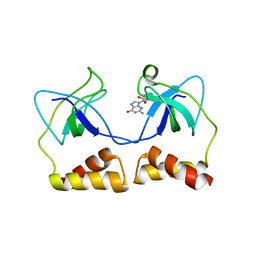 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | DNA (5'-D(*A*G)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWL
 
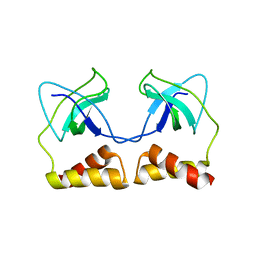 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*(DC))-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWM
 
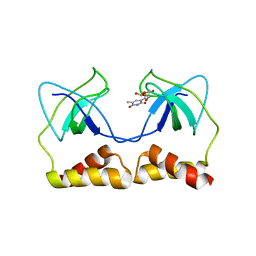 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*T)-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
3W0D
 
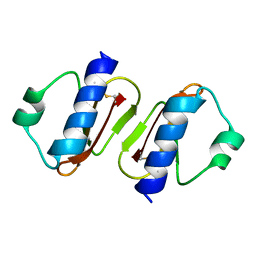 | | Structure of elastase inhibitor AFUEI (cyrstal form I) | | Descriptor: | Elastase inhibitor AFUEI | | Authors: | Imada, K, Sakuma, M, Okumura, Y, Ogawa, K, Nikai, T, Homma, M. | | Deposit date: | 2012-10-29 | | Release date: | 2013-05-15 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Structure Analysis and Characterization of AFUEI, an Elastase Inhibitor from Aspergillus fumigatus
J.Biol.Chem., 288, 2013
|
|
3W0E
 
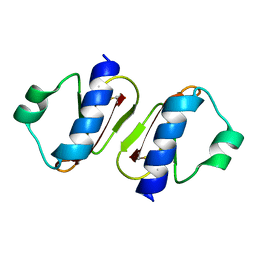 | | Structure of elastase inhibitor AFUEI (crystal form II) | | Descriptor: | Elastase inhibitor AFUEI | | Authors: | Imada, K, Sakuma, M, Okumura, Y, Ogawa, K, Nikai, T, Homma, M. | | Deposit date: | 2012-10-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structure Analysis and Characterization of AFUEI, an Elastase Inhibitor from Aspergillus fumigatus
J.Biol.Chem., 288, 2013
|
|
6G1Q
 
 | | ADP-ribosylserine hydrolase ARH3 of Latimeria chalumnae in complex with ADP-ribose | | Descriptor: | ADP-ribosylhydrolase like 2, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ariza, A. | | Deposit date: | 2018-03-21 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | (ADP-ribosyl)hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition.
Cell Chem Biol, 25, 2018
|
|
