5YL8
 
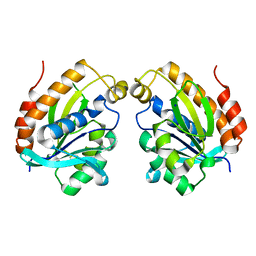 | | The crystal structure of inactive dimeric peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.79 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase, SODIUM ION | | Authors: | Bairagya, H.R, Sharma, P, Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The crystal structure of inactive dimeric peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.79 A resolution
To Be Published
|
|
5YN4
 
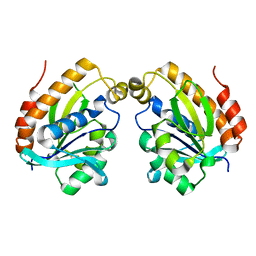 | |
7DAO
 
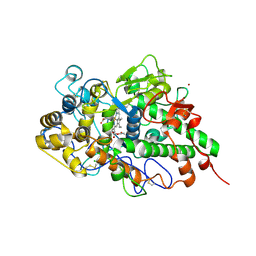 | | Crystal structure of native yak lactoperoxidase at 2.28 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Ahmad, N, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-10-16 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of native yak lactoperoxidase at 2.28 A resolution
To Be Published
|
|
7DY5
 
 | | Structure of the ternary complex of peptidoglycan recognition protein-short (PGRP-S) with hexanoic acid and tartaric acid at 2.30A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Maurya, A, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-01-20 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the ternary complex of peptidoglycan recognition protein-short (PGRP-S)
To Be Published
|
|
7ENU
 
 | | Crystal structure of iron-saturated C-terminal half of lactoferrin produced proteolytically using pepsin at 2.32A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, J, Maurya, A, Viswanathan, V, Singh, P.K, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Crystal structure of iron-saturated C-terminal half of lactoferrin produced proteolytically using pepsin at 2.32A resolution
To Be Published
|
|
8JIV
 
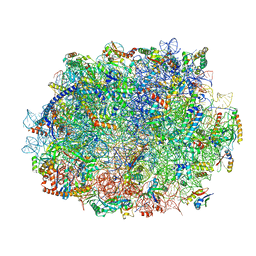 | |
8FTJ
 
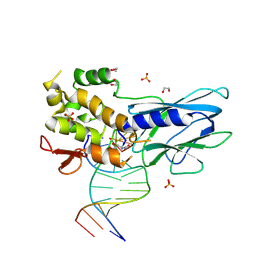 | | Crystal structure of human NEIL1 (P2G (242K) C(delta)100) glycosylase bound to DNA duplex containing urea | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*TP*CP*CP*AP*UDV*GP*TP*CP*TP*AP*CP)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*GP*G)-3'), ... | | Authors: | Tomar, R, Sharma, P, Harp, J.M, Egli, M, Stone, M.P. | | Deposit date: | 2023-01-12 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Base excision repair of the N-(2-deoxy-d-erythro-pentofuranosyl)-urea lesion by the hNEIL1 glycosylase.
Nucleic Acids Res., 51, 2023
|
|
4WF4
 
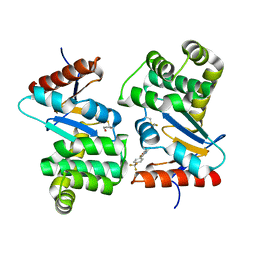 | | Crystal structure of E.Coli DsbA co-crystallised in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-5-carboxylic acid, Thiol:disulfide interchange protein | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7WGK
 
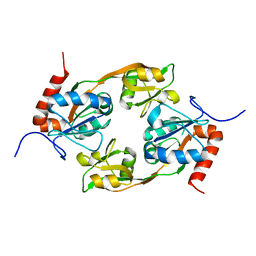 | | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 3.13 A resolution | | Descriptor: | ATP phosphoribosyltransferase | | Authors: | Ahmad, N, Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Singh, T.P, Sharma, S. | | Deposit date: | 2021-12-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 3.13 A resolution
To Be Published
|
|
7WGM
 
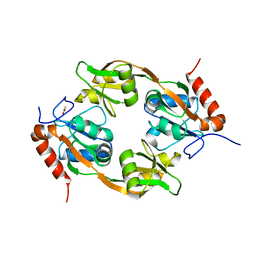 | | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 3.15 A resolution | | Descriptor: | ACETATE ION, ATP phosphoribosyltransferase | | Authors: | Ahmad, N, Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Singh, T.P, Sharma, S. | | Deposit date: | 2021-12-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 3.15 A resolution
To Be Published
|
|
6IDM
 
 | | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Bairagya, H.R, Shokeen, A, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2018-09-10 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution
To Be Published
|
|
6J93
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2019-01-21 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of Peptidyl-tRNA hydrolase form apo at 0.95 A resolution.
To Be Published
|
|
6JQT
 
 | |
6JKX
 
 | | Crystal structure of peptidyl-tRNA hydrolase with multiple sodium and chloride ions at 1.08 A resolution. | | Descriptor: | CHLORIDE ION, METHANOL, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2019-03-03 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase with multiple sodium and chloride ions at 1.08 A resolution.
To Be Published
|
|
6JJ1
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.97 A resolution with disordered five N-terminal residues | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase | | Authors: | Iqbal, N, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2019-02-24 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.97 A resolution with disordered five N-terminal residues
To Be Published
|
|
6JGU
 
 | |
6JJQ
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.99 A resolution. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Bairagya, H.R, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.99 A resolution.
To Be Published
|
|
6KL8
 
 | | Crystal structure of Piptidyl t-RNA hydrolase from Acinetobacter baumannii with bound NaCl at the substrate binding site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of Piptidyl t-RNA hydrolase from Acinetobacter baumannii with bound NaCl at the substrate binding site
To Be Published
|
|
6L2J
 
 | | Crystal structure of yak lactoperoxidase at 1.93 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Sharma, P, Rani, C, Sharma, S, Singh, T.P. | | Deposit date: | 2019-10-04 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Crystal structure of yak lactoperoxidase at 1.93 A resolution.
To Be Published
|
|
5VPN
 
 | | E. coli Quinol fumarate reductase FrdA E245Q mutation | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Starbird, C.A, Maklashina, E, Sharma, P, Qualls-Histed, S, Cecchini, G, Iverson, T.M. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2232 Å) | | Cite: | Structural and biochemical analyses reveal insights into covalent flavinylation of the Escherichia coli Complex II homolog quinol:fumarate reductase.
J. Biol. Chem., 292, 2017
|
|
6KY7
 
 | | Crystal structure of yak lactoperoxidase at 2.27 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Viswanathan, V, Sharma, P, Rani, C, Sharma, S, Singh, T.P. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of yak lactoperoxidase at 2.27 A resolution
To Be Published
|
|
6LSP
 
 | |
6LQW
 
 | | Crystal structure of a dimeric yak lactoperoxidase at 2.59 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Viswanathan, V, Pandey, S.N, Ahmad, N, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-01-14 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a dimeric yak lactoperoxidase at 2.59 A resolution.
To Be Published
|
|
7WZY
 
 | | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 2.975 A resolution | | Descriptor: | ATP phosphoribosyltransferase, FORMIC ACID, GLYCEROL | | Authors: | Ahmad, N, Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.975 Å) | | Cite: | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 2.975 A resolution
To Be Published
|
|
7XFY
 
 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution.
To Be Published
|
|
